Genomic and evolutionary evidence for drought adaptation of allopolyploid Brachypodium hybridum
- PMID: 40102696
- PMCID: PMC12223499
- DOI: 10.1093/jxb/eraf128
Genomic and evolutionary evidence for drought adaptation of allopolyploid Brachypodium hybridum
Abstract
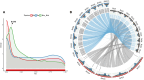
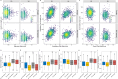
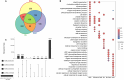
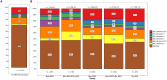
Climate change is increasing the frequency and severity of drought worldwide, threatening the environmental resilience of cultivated grasses. However, the genetic diversity in many wild grasses could contribute to the development of climate-adapted varieties. Here, we elucidated the impact of polyploidy on drought responses using allotetraploid Brachypodium hybridum (B. hybridum) and its progenitor diploid species Brachypodium stacei (B. stacei). Our findings suggest that progenitor species' genomic legacies resulting from hybridization and whole-genome duplications conferred greater ecological adaptive advantages to B. hybridum compared with B. stacei. Genes related to stomatal regulation and the immune response from S-subgenomes were under positive selection during speciation, underscoring their evolutionary importance in adapting to environmental stresses. Biased expression in polyploid subgenomes (B. stacei-type and B. distachyon-type) significantly influenced differential gene expression, with the dominant subgenome exhibiting more differential expression. B. hybridum adapted a drought escape strategy characterized by higher photosynthetic capacity and lower intrinsic water-use efficiency than B. stacei, driven by a highly correlated coexpression network involving genes in the circadian rhythm pathway. In summary, our study shows the influence of polyploidy on ecological and environmental adaptation and resilience in model Brachypodium grasses. These insights hold promise for informing the breeding of climate-resilient cereal crops and pasture grasses.
Keywords: Brachypodium hybridum; Brachypodium stacei; drought response strategy; genomics; polyploidy; stomatal regulation; transcriptomics.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this manuscript.
Figures



Similar articles
-
Variation in functional responses to water stress and differentiation between natural allopolyploid populations in the Brachypodium distachyon species complex.Ann Bot. 2018 Jun 8;121(7):1369-1382. doi: 10.1093/aob/mcy037. Ann Bot. 2018. PMID: 29893879 Free PMC article.
-
Multiple founder events explain the genetic diversity and structure of the model allopolyploid grass Brachypodium hybridum in the Iberian Peninsula hotspot.Ann Bot. 2020 Mar 29;125(4):625-638. doi: 10.1093/aob/mcz169. Ann Bot. 2020. PMID: 31630169 Free PMC article.
-
Environmental niche variation and evolutionary diversification of the Brachypodium distachyon grass complex species in their native circum-Mediterranean range.Am J Bot. 2015 Jul;102(7):1073-88. doi: 10.3732/ajb.1500128. Epub 2015 Jul 16. Am J Bot. 2015. PMID: 26199365
-
Advancements in Water-Saving Strategies and Crop Adaptation to Drought: A Comprehensive Review.Physiol Plant. 2025 Jul-Aug;177(4):e70332. doi: 10.1111/ppl.70332. Physiol Plant. 2025. PMID: 40599019 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Abdoli M, Moieni A, Naghdi Badi H.. 2013. Morphological, physiological, cytological and phytochemical studies in diploid and colchicine-induced tetraploid plants of Echinacea purpurea (L.). Acta Physiologiae Plantarum 35, 2075–2083.
-
- Adem GD, Chen G, Shabala L, Chen Z-H, Shabala S.. 2020. GORK channel: a master switch of plant metabolism? Trends in Plant Science 25, 434–445. - PubMed
-
- Bai X, Shen W, Wu X, Wang P.. 2020. Applicability of long-term satellite-based precipitation products for drought indices considering global warming. Journal of Environmental Management 255, 109846. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

