Single nucleotide polymorphism-based analysis of linkage disequilibrium and runs of homozygosity patterns of indigenous sheep in the southern Taklamakan desert
- PMID: 40102738
- PMCID: PMC11917010
- DOI: 10.1186/s12864-025-11445-9
Single nucleotide polymorphism-based analysis of linkage disequilibrium and runs of homozygosity patterns of indigenous sheep in the southern Taklamakan desert
Abstract
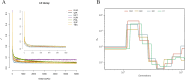
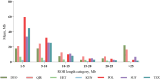
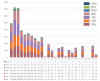
Runs of Homozygosity (ROH) are homozygous genomic fragments inherited from parents to offspring. ROH can be used to indicate the level of inbreeding, as well as to identify possible signatures of artificial or natural selection. Indigenous sheep populations on the southern edge of the Taklimakan Desert have evolved unique genetic traits adapted to extreme desert environments. In an attempt to better understand the adaptive mechanisms of these populations under harsh conditions, we used Illumina® Ovine SNP50K BeadChip to perform a genomic characterization of three recognized breeds (Duolang: n = 36, Hetian: n = 84, Qira black: n = 189) and one ecotypic breed (Kunlun: n = 27) in the region. Additionally, we assessed genomic inbreeding coefficients through ROH analysis, revealing insights into the inbreeding history of these populations. Subsequently, we retrieved candidate genes associated with economic traits in sheep from ROH islands in each breed. To better understand the autozygosity and distribution of ROH islands in these indigenous sheep breeds relative to international breeds, we also included three commercial mutton breeds (Poll Dorset: n = 108, Suffolk: n = 163, Texel: n = 150). The study revealed that among seven sheep breeds, Hetian exhibited the shortest linkage disequilibrium (LD) decay distance, while Kunlun demonstrated the highest LD levels. A total of 10,916 ROHs were obtained. The number of ROHs per breed ranged from 34 (Kunlun) to 2,826 (Texel). The length of ROH was mainly 1-5 Mb (63.54%). Furthermore, 991 candidate genes specific to indigenous sheep breeds were identified, including those associated with heat tolerance, adaptability, energy metabolism, reproduction, and immune response. These findings elucidate the genetic adaptation of indigenous sheep in the Taklimakan Desert, uncovering distinctive characteristics of indigenous sheep formation, and advocating for the conservation and genetic enhancement of local sheep populations.
Keywords: Indigenous sheep; Linkage disequilibrium; ROH islands; Runs of homozygosity; Taklamakan desert.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This work was conducted in accordance with the standards set by the Ethics Committee of of the College of Animal Science and Technology of Tarim University (SYXK2020-009) and approved by the Ethics Committee of of the College of Animal Science and Technology of Tarim University. Written informed consent was obtained from the owners for the participation of their animals in this study. All methods are reported in accordance with ARRIVE guidelines ( https://arriveguidelines.org/ ) for the reporting of animal experiments. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Benefit sharing: The authors declare no benefit sharing.
Figures

Similar articles
-
Population structure and selection signal analysis of indigenous sheep from the southern edge of the Taklamakan Desert.BMC Genomics. 2024 Jul 9;25(1):681. doi: 10.1186/s12864-024-10581-y. BMC Genomics. 2024. PMID: 38982349 Free PMC article.
-
Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data.BMC Genomics. 2021 Jan 6;22(1):7. doi: 10.1186/s12864-020-07314-2. BMC Genomics. 2021. PMID: 33407115 Free PMC article.
-
Analysis on the desert adaptability of indigenous sheep in the southern edge of Taklimakan Desert.Sci Rep. 2022 Jul 18;12(1):12264. doi: 10.1038/s41598-022-15986-x. Sci Rep. 2022. PMID: 35851076 Free PMC article.
-
Runs of homozygosity: current knowledge and applications in livestock.Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review.
-
Genomic advancements in goat breeding: enhancing productivity, disease resistance, and sustainability in India's rural economy.Mamm Genome. 2025 May 28. doi: 10.1007/s00335-025-10138-8. Online ahead of print. Mamm Genome. 2025. PMID: 40434651 Review.
References
-
- Yang F, He Q, Huang J, Mamtimin A, Yang X, Huo W, Zhou C, Liu X, Wei W, Cui C, Wang M, Li H, Yang L, Zhang H, Liu Y, Zheng X, Pan H, Jin L, Zou H, Zhou L, Liu Y, Zhang J, Meng L, Wang Y, Qin X, Yao Y, Liu H, Xue F, Zheng W. Desert environment and climate observation network over the Taklimakan desert. Bull Am Meteorol Soc. 2020;102(6):E1172–91. 10.1175/BAMS-D-20-0236.1
-
- Xu X, Wang Y, Wei W, Zhao T, Xu X. Summertime precipitation process and atmospheric water cycle over Tarim basin under the specific large terrain background. Desert Oasis Meteorol. 2014;2014(82):1–11. 10.3969/j.issn.1002-0799.2014.02.001
-
- Gui Dongwei Z, Fanjiang L, Zhen Z Bo. Characteristics of the clonal propagation of Alhagi sparsifolia Shap. (Fabaceae) under different groundwater depths in Xinjiang, China. Rangel J. 2013;35:355–62. 10.1071/RJ13004
-
- Ma M, Yang X, He Q, Zhou C, Yang F. Characteristics of dust devil and its dust emission in Northern margin of the Taklimakan desert. Aeolian Res. 2020;44:100579. 10.1016/j.aeolia.2020.100579
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

