An mRNA expression atlas for the duck with public RNA-seq datasets
- PMID: 40102741
- PMCID: PMC11916966
- DOI: 10.1186/s12864-025-11385-4
An mRNA expression atlas for the duck with public RNA-seq datasets
Abstract
Background: Ducks are globally important poultry species and a major source of farm animal products, including meat, eggs, and feathers. A thorough understanding of the functional genomic and transcriptomic sequences is crucial for improving production efficiency.
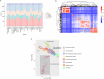
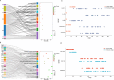
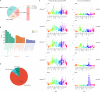
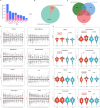
Result: This study constructed the largest duck mRNA expression atlas among all waterfowl species to date. The atlas encompasses 1,257 tissue samples across 30 tissue types, representing all major organ systems. Using advanced clustering analysis, we established co-expression network clusters to describe the transcriptional features in the duck mRNA expression atlas and, when feasible, assign these features to unique tissue types or pathways. Additionally, we identified 27 low-variance, highly expressed housekeeping genes suitable for gene expression experiments. Furthermore, in-depth analysis revealed potential sex-biased gene expression patterns within tissues and specific gene expression profiles in meat-type and egg-type ducks, providing valuable resources to understand the genetic basis of sex differences and particular phenotypes. This research elucidates the biological processes affecting duck productivity.
Conclusion: This study presents the most extensive gene expression atlas for any waterfowl species to date. These findings are of significant value for advancing duck biological research and industrial applications.
Keywords: Co-expression network; Differentially expressed genes; Duck; Expression atlas; Housekeeping genes; RNA-seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All methods were carried out following relevant guidelines and regulations. All duck work was conducted following a protocol approved by Sichuan Agricultural University China’s animal ethics and welfare committee (AEWC). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Li J. Analysis of the evolution of sex chromosomes in Pekin Duck and monotremes. Zhejiang University; 2020.
-
- Wickramasinghe S, Cánovas A, Rincón G, Medrano JF. RNA-sequencing: a tool to explore new frontiers in animal genetics. Livest Sci. 2014;166:206–16. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

