Integrated bioinformatics and validation reveal TMEM45A in systemic lupus erythematosus regulating atrial fibrosis in atrial fibrillation
- PMID: 40102753
- PMCID: PMC11917082
- DOI: 10.1186/s10020-025-01162-0
Integrated bioinformatics and validation reveal TMEM45A in systemic lupus erythematosus regulating atrial fibrosis in atrial fibrillation
Abstract
Background: Accumulative evidence has shown that systemic lupus erythematosus (SLE) increases the risk of various cardiovascular diseases including atrial fibrillation (AF). The study aimed to screen potential key genes underlying co-pathogenesis between SLE and AF, and to discover therapeutic targets for AF.
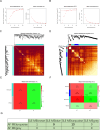
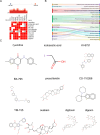
Methods: Differentially expressed genes (DEGs) were identified, and co-expressed gene modules were obtained through weighted gene co-expression network analysis (WGCNA) based on the AF and SLE expression profiles from the GEO database. Subsequently, machine learning algorithms including LASSO regression and support vector machine (SVM) method were employed to identify the candidate therapeutic target for SLE-related AF. Furthermore, the therapeutic role of TMEM45A was validated both in vivo and vitro.
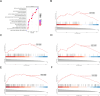
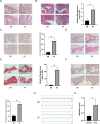
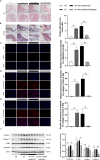
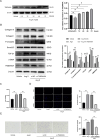
Results: Totally, 26 DEGs were identified in SLE and AF. The PPI network combined with WGCNA identified 51 key genes in SLE and AF. Ultimately, Machine learning-based methods screened three hub genes in SLE combined with AF, including TMEM45A, ITGB2 and NFKBIA. The cMAP analysis exposed KI-8751 and YM-155 as potential drugs for AF treatment. Regarding TMEM45A, the aberrant expression was validated in blood of SLE patients. Additionally, TMEM45A expression was up-regulated in the atrial tissue of patients with AF. Furthermore, TMEM45A knockdown alleviated AF occurrence and atrial fibrosis in vivo and Ang II-induced NRCFs fibrosis in vitro.
Conclusion: The crosstalk genes underlying co-pathogenesis between SLE and AF were unraveled. Furthermore, the pro-fibrotic role of TMEM45A was validated in vivo and vitro, highlighting its potential as a therapeutic target for AF.
Keywords: Atrial fibrillation; Bioinformatics; Fibrosis; Systemic lupus erythematosus; TMEM45A.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Ethics Committee of Changhai Hospital, Naval Medical University. We confirmed that all experiments were performed in accordance with the regulations. Consent for publication: All authors confirm their consent for publication the manuscript. Competing interests: The authors declare no competing interests.
Figures





References
-
- Apte M, et al. Associated factors and impact of myocarditis in patients with SLE from LUMINA, a multiethnic US cohort. Rheumatology. 2008;47:362–7. - PubMed
-
- Brundel B, et al. Atrial fibrillation. Nat Rev Dis Primers. 2022;8:21. - PubMed
-
- Chang JH, et al. Vascular endothelial growth factor modulates pulmonary vein arrhythmogenesis via vascular endothelial growth factor receptor 1/NOS pathway. Eur J Pharmacol. 2021;911:174547. - PubMed
-
- Chen Y, Fu L, Pu S, Xue Y. Systemic lupus erythematosus increases risk of incident atrial fibrillation: a systematic review and meta-analysis. Int J Rheum Dis. 2022;25:1097–106. - PubMed
-
- Gorji AE, Roudbari Z, Alizadeh A, Sadeghi B. Investigation of systemic lupus erythematosus (SLE) with integrating transcriptomics and genome wide association information. Gene. 2019;706:181–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

