Lactate accumulation induces H4K12la to activate super-enhancer-driven RAD23A expression and promote niraparib resistance in ovarian cancer
- PMID: 40102876
- PMCID: PMC11921584
- DOI: 10.1186/s12943-025-02295-w
Lactate accumulation induces H4K12la to activate super-enhancer-driven RAD23A expression and promote niraparib resistance in ovarian cancer
Abstract
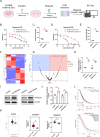
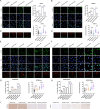
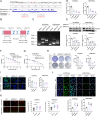
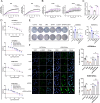
Ovarian cancer is a gynecological malignancy with the highest recurrence and mortality rates. Although niraparib can effectively affect its progression, the challenge of drug resistance remains. Herein, niraparib-resistant ovarian cancer cell lines were constructed to identify the abnormally activated enhancers and associated target genes via RNA in situ conformation sequencing. Notably, the target gene RAD23A was markedly upregulated in niraparib-resistant cells, and inhibiting RAD23A restored their sensitivity. Additionally, abnormal activation of glycolysis in niraparib-resistant cells induced lactate accumulation, which promoted the lactylation of histone H4K12 lysine residues. Correlation analysis showed that key glycolysis enzymes such as pyruvate kinase M and lactate dehydrogenase A were significantly positively correlated with RAD23A expression in ovarian cancer. Additionally, H4K12la activated the super-enhancer (SE) of niraparib and RAD23A expression via MYC transcription factor, thereby enhancing the DNA damage repair ability and promoting the drug resistance of ovarian cancer cells. Overall, the findings of this study indicate that lactic acid accumulation leads to lactylation of histone H4K12la, thereby upregulating SE-mediated abnormal RAD23A expression and promoting niraparib resistance in ovarian cancer cells, suggesting RAD23A as a potential therapeutic target for niraparib-resistant ovarian cancer.
Keywords: Glycolysis; H4K12la; Niraparib resistance; RAD23A; Super-enhancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Ethics Committee of the third affiliated Hospital of Guangzhou Medical University [NO: 2024 − 301] and Liaoning Cancer Hospital & Institute [NO: KY20231103] approved the use of human tissue in this study. Informed consent was obtained from all patients. Animal experiments were approved by the Experimental Animal Ethics Committee of Guangzhou Medical University [NO: S2024-004]. Consent for publication: All authors have reviewed the final version of the manuscript and approve it for publication. Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

