Analysis of 30 chromosome-level Drosophila genome assemblies reveals dynamic evolution of centromeric satellite repeats
- PMID: 40102968
- PMCID: PMC11917152
- DOI: 10.1186/s13059-025-03527-4
Analysis of 30 chromosome-level Drosophila genome assemblies reveals dynamic evolution of centromeric satellite repeats
Abstract
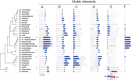
Background: The Drosophila genus is ideal for studying genome evolution due to its relatively simple chromosome structure and small genome size, with rearrangements mainly restricted to within chromosome arms, such as Muller elements. However, work on the rapidly evolving repetitive genomic regions, composed of transposons and tandem repeats, have been hampered by the lack of genus-wide chromosome-level assemblies.
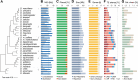
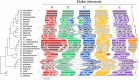
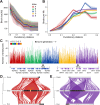
Results: Integrating long-read genomic sequencing and chromosome capture technology, here we produce and annotate 30 chromosome-level genome assemblies within the Drosophila genus. Based on this dataset, we reveal the evolutionary dynamics of genome rearrangements across the Drosophila phylogeny, including the identification of genomic regions that show comparatively high structural stability throughout evolution. Moreover, within the ananassae subgroup, we uncover the emergence of new chromosome conformations and the rapid expansion of novel satellite DNA sequence families, which form large and continuous pericentromeric domains with higher-order repeat structures that are reminiscent of those observed in the human and Arabidopsis genomes.
Conclusions: These chromosome-level genome assemblies present a valuable resource for future research, the power of which is demonstrated by our analysis of genome rearrangements and chromosome evolution. In addition, based on our findings, we propose the ananassae subgroup as an ideal model system for studying the evolution of centromere structure.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures



Similar articles
-
De novo identification of satellite DNAs in the sequenced genomes of Drosophila virilis and D. americana using the RepeatExplorer and TAREAN pipelines.PLoS One. 2019 Dec 19;14(12):e0223466. doi: 10.1371/journal.pone.0223466. eCollection 2019. PLoS One. 2019. PMID: 31856171 Free PMC article.
-
High-Quality Genome Assemblies Reveal Evolutionary Dynamics of Repetitive DNA and Structural Rearrangements in the Drosophila virilis Subgroup.Genome Biol Evol. 2024 Jan 5;16(1):evad238. doi: 10.1093/gbe/evad238. Genome Biol Evol. 2024. PMID: 38159044 Free PMC article.
-
Four near-complete genome assemblies reveal the landscape and evolution of centromeres in Salicaceae.Genome Biol. 2025 May 2;26(1):111. doi: 10.1186/s13059-025-03578-7. Genome Biol. 2025. PMID: 40317068 Free PMC article.
-
Sequence, Chromatin and Evolution of Satellite DNA.Int J Mol Sci. 2021 Apr 21;22(9):4309. doi: 10.3390/ijms22094309. Int J Mol Sci. 2021. PMID: 33919233 Free PMC article. Review.
-
Helitrons shaping the genomic architecture of Drosophila: enrichment of DINE-TR1 in α- and β-heterochromatin, satellite DNA emergence, and piRNA expression.Chromosome Res. 2015 Sep;23(3):597-613. doi: 10.1007/s10577-015-9480-x. Chromosome Res. 2015. PMID: 26408292 Review.
References
-
- Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J Exp Zool. 1913;14:43–59.
-
- Bridges CB. SALIVARY CHROMOSOME MAPS: with a key to the banding of the chromosomes of Drosophila Melanogaster. J Hered. 1935;26:60–4.
-
- Dobzhansky T. Genetics and the origin of species. New York: Columbia University Press; 1937.
-
- Muller HJ. An analysis of the process of structural change in chromosomes of Drosophila. J Genet. 1940;40:1–66.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

