SARS-CoV2 infection triggers inflammatory conditions and astrogliosis-related gene expression in long-term human cortical organoids
- PMID: 40103011
- PMCID: PMC12121356
- DOI: 10.1093/stmcls/sxaf010
SARS-CoV2 infection triggers inflammatory conditions and astrogliosis-related gene expression in long-term human cortical organoids
Abstract
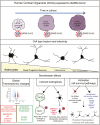
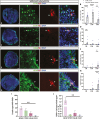
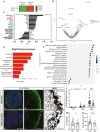
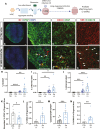
SARS-CoV2, severe acute respiratory syndrome coronavirus 2, is frequently associated with neurological manifestations. Despite the presence of mild to severe CNS-related symptoms in a cohort of patients, there is no consensus whether the virus can infect directly brain tissue or if the symptoms in patients are a consequence of peripheral infectivity of the virus. Here, we use long-term human stem cell-derived cortical organoids to assess SARS-CoV2 infectivity of brain cells and unravel the cell-type tropism and its downstream pathological effects. Our results show consistent and reproducible low levels of SARS-CoV2 infection of astrocytes, deep projection neurons, upper callosal neurons, and inhibitory neurons in 6 months of human cortical organoids. Interestingly, astrocytes showed the highest infection rate among all infected cell populations which led to changes in their morphology and upregulation of SERPINA3, CD44, and S100A10 astrogliosis markers. Further, transcriptomic analysis revealed overall changes in expression of genes related to cell metabolism, astrogliosis and, inflammation and further, upregulation of cell survival pathways. Thus, local and minor infectivity of SARS-CoV2 in the brain may induce widespread adverse effects and lead to the resilience of dysregulated neurons and astrocytes within an inflammatory environment.
Keywords: SARS-CoV2 brain infectivity; astrogliosis; human cortical organoids; inflammatory and cell survival mechanisms.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Zhu N, Zhang D, Wang W, et al.; China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. https://doi.org/10.1056/NEJMoa2001017 - DOI - PMC - PubMed
-
- Kuba K, Imai Y, Rao S, et al.A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875-879. https://doi.org/10.1038/nm1267 - DOI - PMC - PubMed
-
- Ding Y, He L, Zhang Q, et al.Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203:622-630. https://doi.org/10.1002/path.1560 - DOI - PMC - PubMed
-
- Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al.Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590-592. https://doi.org/10.1056/NEJMc2011400 - DOI - PMC - PubMed
-
- Mao L, Jin H, Wang M, et al.Neurologic manifestations of hospitalized patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683-690. https://doi.org/10.1001/jamaneurol.2020.1127 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

