VirBR counter-silences HppX3 to promote conjugation of blaNDM-IncX3 plasmids
- PMID: 40103225
- PMCID: PMC11915502
- DOI: 10.1093/nar/gkaf182
VirBR counter-silences HppX3 to promote conjugation of blaNDM-IncX3 plasmids
Abstract
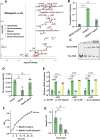
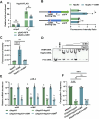
New Delhi metallo-β-lactamases (NDM), encoded by the blaNDM gene, mediate carbapenem resistance, posing serious threats to public health due to their global presence across diverse hosts and environments. The blaNDM is prominently carried by the IncX3 plasmid, which encodes a Type IV secretion system (T4SS) responsible for plasmid conjugation. This T4SS has been shown to be phenotypically silenced by a plasmid-borne H-NS family protein; however, the underlying mechanisms of both silencing and silencing relief remain unclear. Herein, we identified HppX3, an H-NS family protein encoded by the IncX3 plasmid, as a transcription repressor. HppX3 binds to the T4SS promoter (PactX), downregulates T4SS expression, thereby inhibits plasmid conjugation. RNA-seq analysis revealed that T4SS genes are co-regulated by HppX3 and VirBR, a transcription activator encoded by the same plasmid. Mechanistically, VirBR acts as a counter-silencer by displacing HppX3 from PactX, restoring T4SS expression and promoting plasmid conjugation. A similar counter-silencing mechanism was identified in the T4SSs of IncX1 and IncX2 plasmids. These findings provide new insights into the regulatory mechanisms controlling T4SS expression on multiple IncX plasmids, including the IncX3, explaining the persistence and widespread of blaNDM-IncX3 plasmid, and highlight potential strategies to combat the spread of NDM-positive Enterobacterales by targeting plasmid-encoded regulators.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures




References
-
- Yong D, Toleman MA, Giske CG et al. . Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009; 53:5046–54.10.1128/AAC.00774-09. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

