m6A2Circ: A comprehensive database for decoding the regulatory relationship between m6A modification and circular RNA
- PMID: 40103610
- PMCID: PMC11914901
- DOI: 10.1016/j.csbj.2025.02.018
m6A2Circ: A comprehensive database for decoding the regulatory relationship between m6A modification and circular RNA
Abstract
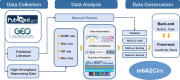
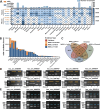
Circular RNA (circRNA) is a class of noncoding RNAs derived from back-splicing of pre-mRNAs. Recent studies have increasingly highlighted the pivotal roles of N6-methyladenosine (m6A) in regulating various aspects of circRNA metabolism, including biogenesis, localization, stability, and translation. Despite the importance of m6A in circRNA metabolism, there remains a substantial gap in comprehensive resources dedicated to exploring m6A modification in circRNA. To bridge this significant gap, we present m6A2Circ (http://m6a2circ.canceromics.org/), a pioneering database designed to systematically explore the regulatory interactions between m6A modification and circRNA. The m6A2Circ database encompasses 198,804 m6A-circRNA associations derived from diverse human and mouse tissues. These associations are meticulously categorized into four levels of evidence supported either by experimental data or by high-throughput sequencing data. Moreover, the database offers extensive annotations, facilitating research into circRNA function and its potential disease implications. Overall, m6A2Circ aims to benefit the research community and bolster novel discoveries in terms of crosstalk between m6A and circRNA.
Keywords: Circular RNA; Database; N6-methyladenosine.
© 2025 Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
N6-Methyladenosine Modification Opens a New Chapter in Circular RNA Biology.Front Cell Dev Biol. 2021 Jul 23;9:709299. doi: 10.3389/fcell.2021.709299. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34368159 Free PMC article. Review.
-
Novel insights into the N 6-methyladenosine modification on circRNA in cancer.Front Oncol. 2025 May 19;15:1554888. doi: 10.3389/fonc.2025.1554888. eCollection 2025. Front Oncol. 2025. PMID: 40458727 Free PMC article. Review.
-
The functional roles, cross-talk and clinical implications of m6A modification and circRNA in hepatocellular carcinoma.Int J Biol Sci. 2021 Jul 22;17(12):3059-3079. doi: 10.7150/ijbs.62767. eCollection 2021. Int J Biol Sci. 2021. PMID: 34421350 Free PMC article. Review.
-
Recent advances in crosstalk between N6-methyladenosine (m6A) modification and circular RNAs in cancer.Mol Ther Nucleic Acids. 2022 Jan 21;27:947-955. doi: 10.1016/j.omtn.2022.01.013. eCollection 2022 Mar 8. Mol Ther Nucleic Acids. 2022. PMID: 35211355 Free PMC article. Review.
-
Crosstalk between m6A mRNAs and m6A circRNAs and the time-specific biogenesis of m6A circRNAs after OGD/R in primary neurons.Epigenetics. 2023 Dec;18(1):2181575. doi: 10.1080/15592294.2023.2181575. Epigenetics. 2023. PMID: 36861189 Free PMC article.
References
-
- Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H., et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019;177:865–880. e821. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
