A general approach for activity-based protein profiling of oxidoreductases with redox-differentiated diarylhalonium warheads
- PMID: 40103729
- PMCID: PMC11912224
- DOI: 10.1039/d4sc08454c
A general approach for activity-based protein profiling of oxidoreductases with redox-differentiated diarylhalonium warheads
Abstract
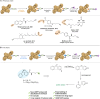
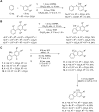
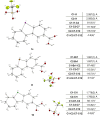
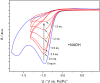
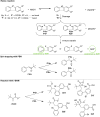
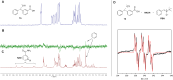
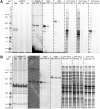
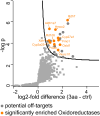
Activity-based protein profiling (ABPP) is a unique proteomic tool for measuring the activity of enzymes in their cellular context, which has been well established for enzyme classes exhibiting a characteristic nucleophilic residue (e.g., hydrolases). In contrast, the enzyme class of oxidoreductases has received less attention, as its members rely mainly on cofactors instead of nucleophilic amino acid residues for catalysis. ABPP probes have been designed for specific oxidoreductase subclasses, which rely on the oxidative conversion of the probes into strong electrophiles. Here we describe the development of ABPP probes for the simultaneous labeling of various subclasses of oxidoreductases. The probe warheads are based on hypervalent diarylhalonium salts, which show unique reactivity as their activation proceeds via a reductive mechanism resulting in aryl radicals leading to covalent labeling of liver proteins at several different amino acids in close proximity to the active sites. The redox potential of the probes can be tuned by isosteric replacement varying the halonium central atom. ABPP experiments with liver using 16 probes differing in warhead, linker, and structure revealed distinct overlapping profiles and broad substrate specificities of several probes. With their capability of multi oxidoreductase subclass labeling - including rare examples for the class of reductases - and their unique design, the herein reported probes offer new opportunities for the investigation of the "oxidoreductome" of microorganisms, plants, animal and human tissues.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
[Advances in applications of activity-based chemical probes in the characterization of amino acid reactivities].Se Pu. 2023 Jan;41(1):14-23. doi: 10.3724/SP.J.1123.2022.05013. Se Pu. 2023. PMID: 36633073 Free PMC article. Review. Chinese.
-
Activity-Based Protein Profiling (ABPP) of Oxidoreductases.Chembiochem. 2021 Feb 15;22(4):630-638. doi: 10.1002/cbic.202000542. Epub 2020 Oct 20. Chembiochem. 2021. PMID: 32881211 Free PMC article. Review.
-
Covalent protein modification: the current landscape of residue-specific electrophiles.Curr Opin Chem Biol. 2015 Feb;24:18-26. doi: 10.1016/j.cbpa.2014.10.021. Epub 2014 Nov 11. Curr Opin Chem Biol. 2015. PMID: 25461720 Review.
-
Activity-Based Hydrazine Probes for Protein Profiling of Electrophilic Functionality in Therapeutic Targets.ACS Cent Sci. 2021 Sep 22;7(9):1524-1534. doi: 10.1021/acscentsci.1c00616. Epub 2021 Aug 19. ACS Cent Sci. 2021. PMID: 34584954 Free PMC article.
-
Decaging-to-labeling: Development and investigation of quinone methide warhead for protein labeling.Bioorg Chem. 2024 Feb;143:107088. doi: 10.1016/j.bioorg.2023.107088. Epub 2024 Jan 4. Bioorg Chem. 2024. PMID: 38194902
References
-
- Yang P. Liu K. ChemBioChem. 2015;16:712. - PubMed
- Willems L. I. Overkleeft H. S. van Kasteren S. I. Bioconjugate Chem. 2014;25:1181. - PubMed
- Roberts A. M. Ward C. C. Nomura D. K. Curr. Opin. Biotechnol. 2017;43:25. - PMC - PubMed
- Cravatt B. F. Wright A. T. Kozarich J. W. Annu. Rev. Biochem. 2008;77:383. - PubMed
- Evans M. J. Cravatt B. F. Chem. Rev. 2006;106:3279. - PubMed
- Fang H. Peng B. Ong S. Y. Wu Q. Li L. Yao S. Q. Chem. Sci. 2021;12:8288. doi: 10.1039/D1SC01359A. - DOI - PMC - PubMed
-
- Cichocki B. A. Khobragade V. Donzel M. Cotos L. Blandin S. Schaeffer-Reiss C. Cianférani S. Strub J.-M. Elhabiri M. Davioud-Charvet E. JACS Au. 2021;1:669. - PMC - PubMed
- Simon G. M. Cravatt B. F. J. Biol. Chem. 2010;285:11051. - PMC - PubMed
- Greenbaum D. Medzihradszky K. F. Burlingame A. Bogyo M. Chem. Biol. 2000;7:569. - PubMed
- Kato D. Boatright K. M. Berger A. B. Nazif T. Blum G. Ryan C. Chehade K. A. H. Salvesen G. S. Bogyo M. Nat. Chem. Biol. 2005;1:33. doi: 10.1038/nchembio707. - DOI - PubMed
- Clerc J. Florea B. I. Kraus M. Groll M. Huber R. Bachmann A. S. Dudler R. Driessen C. Overkleeft H. S. Kaiser M. ChemBioChem. 2009;10:2638. doi: 10.1002/cbic.200900411. - DOI - PubMed
- Florea B. I. Verdoes M. Li N. van der Linden W. A. Geurink P. P. van den Elst H. Hofmann T. de Ru A. van Veelen P. A. Tanaka K. Sasaki K. Murata S. den Dulk H. Brouwer J. Ossendorp F. A. Kisselev A. F. Overkleeft H. S. Chem. Biol. 2010;17:795. - PMC - PubMed
- Witte M. D. Walvoort M. T. C. Li K.-Y. Kallemeijn W. W. Donker-Koopman W. E. Boot R. G. Aerts J. M. F. G. Codée J. D. C. van der Marel G. A. Overkleeft H. S. ChemBioChem. 2011;12:1263. doi: 10.1002/cbic.201000773. - DOI - PubMed
-
- Obianyo O. Causey C. P. Jones J. E. Thompson P. R. ACS Chem. Biol. 2011;6:1127. - PMC - PubMed
- Adam G. C. Sorensen E. J. Cravatt B. F. Nat. Biotechnol. 2002;20:805. doi: 10.1038/nbt714. - DOI - PubMed
- Liu Y. Shreder K. R. Gai W. Corral S. Ferris D. K. Rosenblum J. S. Chem. Biol. 2005;12:99. - PubMed
- Liu Y. Jiang N. Wu J. Dai W. Rosenblum J. S. J. Biol. Chem. 2007;282:2505. doi: 10.1074/jbc.M609603200. - DOI - PubMed
- Patricelli M. P. Szardenings A. K. Liyanage M. Nomanbhoy T. K. Wu M. Weissig H. Aban A. Chun D. Tanner S. Kozarich J. W. Biochemistry. 2007;46:350. doi: 10.1021/bi062142x. - DOI - PubMed
- Shi H. Cheng X. Sze S. K. Yao S. Q. Chem. Commun. 2011;47:11306. doi: 10.1039/C1CC14824A. - DOI - PubMed
- Lu C. H. S. Liu K. Tan L. P. Yao S. Q. Chem. - Eur. J. 2012;18:28. - PubMed
-
- Deng H. Lei Q. Wu Y. He Y. Li W. Eur. J. Med. Chem. 2020;191:112151. - PubMed
- Xu J. Li X. Ding K. Li Z. Chem. - Asian J. 2020;15:34. doi: 10.1002/asia.201901500. - DOI - PubMed
- Pichler C. M. Krysiak J. Breinbauer R. Bioorg. Med. Chem. 2016;24:3291. doi: 10.1016/j.bmc.2016.03.050. - DOI - PubMed
- Benns H. J. Wincott C. J. Tate E. W. Child M. A. Curr. Opin. Chem. Biol. 2021;60:20. - PubMed
- Carvalho L. A. R. Bernardes G. J. L. ChemMedChem. 2022;17:e202200174. - PMC - PubMed
LinkOut - more resources
Full Text Sources

