Contribution of intact viral genomes persisting in blood and tissues during ART to plasma viral rebound in SHIV-infected rhesus macaques
- PMID: 40104070
- PMCID: PMC11914814
- DOI: 10.1016/j.isci.2025.111998
Contribution of intact viral genomes persisting in blood and tissues during ART to plasma viral rebound in SHIV-infected rhesus macaques
Abstract
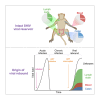
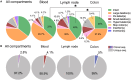
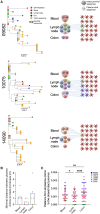
Persistent SIV/HIV reservoirs are the primary obstacle to a cure and the source of viral rebound after ART interruption (ATI). However, the anatomical source of viral rebound remains elusive. Here, we characterized the proviral landscape in the blood, inguinal, and axillary lymph nodes and colon biopsies of five SHIV-infected rhesus macaques (RMs), under ART for 28 weeks. From the 144 near full-length (NFL) proviral sequences obtained pre-ATI, 35% were genetically intact and only 2.8% were found in multiple copies. Envelope sequences of plasma rebounding viruses after ATI, more frequently matched pre-ATI intact proviruses retrieved from lymph nodes compared to sequences isolated from the blood or the colon (4, 1, and 1 pair of matched sequences, respectively). Our results suggest that clonal expansion of infected cells rare in this model, and that intact proviruses persisting in the lymph nodes may be a preferential source of viral rebound upon ATI.
Keywords: Genomics; Virology.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R., et al. Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. - DOI - PubMed
-
- Chun T.-W., Stuyver L., Mizell S.B., Ehler L.A., Mican J.A., Baseler M., Lloyd A.L., Nowak M.A., Fauci A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. - DOI - PMC - PubMed
-
- Rothenberger M.K., Keele B.F., Wietgrefe S.W., Fletcher C.V., Beilman G.J., Chipman J.G., Khoruts A., Estes J.D., Anderson J., Callisto S.P., et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc. Natl. Acad. Sci. USA. 2015;112:E1126–E1134. doi: 10.1073/pnas.1414926112. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

