Cell surface heparan sulfate is an attachment receptor for grass carp reovirus
- PMID: 40104073
- PMCID: PMC11914516
- DOI: 10.1016/j.isci.2025.112033
Cell surface heparan sulfate is an attachment receptor for grass carp reovirus
Abstract
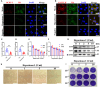
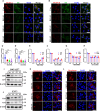
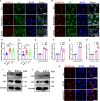
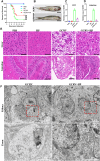
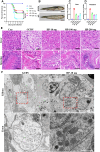
Grass carp reovirus (GCRV) causes hemorrhagic disease in grass carp, leading to significant economic losses in China's aquaculture. However, the cellular receptors responsible for the initiation of GCRV infection remain unclear. This study reveals that cell surface heparan sulfate (HS) acts as a crucial attachment receptor for GCRV. Removing HS with heparinase significantly reduces GCRV attachment and infection. Both HS and its homologue, heparin, inhibit the attachment of GCRV to cells. Altering HS levels in cells affects GCRV attachment and infection accordingly. GCRV outer capsid proteins VP5, VP56, and VP35, as well as purified GCRV virions, directly bind to HS. Pretreating GCRV with heparin or feeding grass carp with feed containing heparin significantly reduces mortality caused by GCRV infection. Collectively, these results highlight the crucial role of HS as an attachment receptor for GCRV and therefore provide a promising target for the prevention and control of this virus.
Keywords: Aquaculture; Aquaculture diseases; Biochemistry; Cell biology; Virology.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Fisheries Bureau of Ministry of Agriculture and Rural Affairs in China . China Agriculture Press; 2023. China Fishery Statistical Yearbook of 2022.
-
- Fang Q., Attoui H., Cantaloube J.F., Biagini P., Zhu Z., de Micco P., de Lamballerie X. Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (Genus Aquareovirus, family Reoviridae) Biochem. Biophys. Res. Commun. 2000;274:762–766. - PubMed
-
- Ye X., Tian Y.Y., Deng G.C., Chi Y.Y., Jiang X.Y. Complete genomic sequence of a reovirus isolated from grass carp in China. Virus Res. 2012;163:275–283. - PubMed
-
- Pei C., Ke F., Chen Z.Y., Zhang Q.Y. Complete genome sequence and comparative analysis of grass carp reovirus strain 109 (GCReV-109) with other grass carp reovirus strains reveals no significant correlation with regional distribution. Arch. Virol. 2014;159:2435–2440. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

