Estimation of ionic currents and compensation mechanisms from recursive piecewise assimilation of electrophysiological data
- PMID: 40104428
- PMCID: PMC11913807
- DOI: 10.3389/fncom.2025.1458878
Estimation of ionic currents and compensation mechanisms from recursive piecewise assimilation of electrophysiological data
Abstract
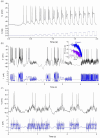
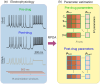
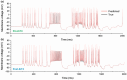
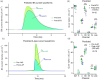
The identification of ion channels expressed in neuronal function and neuronal dynamics is critical to understanding neurological disease. This program calls for advanced parameter estimation methods that infer ion channel properties from the electrical oscillations they induce across the cell membrane. Characterization of the expressed ion channels would allow detecting channelopathies and help devise more effective therapies for neurological and cardiac disease. Here, we describe Recursive Piecewise Data Assimilation (RPDA), as a computational method that successfully deconvolutes the ionic current waveforms of a hippocampal neuron from the assimilation of current-clamp recordings. The strength of this approach is to simultaneously estimate all ionic currents in the cell from a small but high-quality dataset. RPDA allows us to quantify collateral alterations in non-targeted ion channels that demonstrate the potential of the method as a drug toxicity counter-screen. The method is validated by estimating the selectivity and potency of known ion channel inhibitors in agreement with the standard pharmacological assay of inhibitor potency (IC50).
Keywords: data assimilation; dynamical systems; ion channels; neurons and networks; parameter estimation.
Copyright © 2025 Wells, Morris, Taylor and Nogaret.
Conflict of interest statement
The authors declare that part of this research was patented under patent number WO2023/214156.
Figures

Similar articles
-
Single shot detection of alterations across multiple ionic currents from assimilation of cell membrane dynamics.Sci Rep. 2024 Mar 12;14(1):6031. doi: 10.1038/s41598-024-56576-3. Sci Rep. 2024. PMID: 38472404 Free PMC article.
-
Non-synaptic ion channels in insects--basic properties of currents and their modulation in neurons and skeletal muscles.Prog Neurobiol. 2001 Aug;64(5):431-525. doi: 10.1016/s0301-0082(00)00066-6. Prog Neurobiol. 2001. PMID: 11301158 Review.
-
A- and C-type rat nodose sensory neurons: model interpretations of dynamic discharge characteristics.J Neurophysiol. 1994 Jun;71(6):2338-58. doi: 10.1152/jn.1994.71.6.2338. J Neurophysiol. 1994. PMID: 7523613
-
Parameter estimation in cardiac ionic models.Prog Biophys Mol Biol. 2004 Jun-Jul;85(2-3):407-31. doi: 10.1016/j.pbiomolbio.2004.02.002. Prog Biophys Mol Biol. 2004. PMID: 15142755
-
A Short Guide to Electrophysiology and Ion Channels.J Pharm Pharm Sci. 2017;20:48-67. doi: 10.18433/J32P6R. J Pharm Pharm Sci. 2017. PMID: 28459656 Review.
References
-
- Abarbanel H. D. I. (2022). The statistical physics of data assimilation and machine learning. Cambridge: CUP.
-
- Abarbanel H. D. I., Kostuk M., Whartenby W. (2010). Data assimilation with regularized nonlinear instabilities. Q. J. R. Meteorol. Soc. 136, 769–783. doi: 10.1002/qj.600 - DOI
-
- Ahrens M., Paninski L., Huys Q. (2005). “Large-scale biophysical parameter estimation in single neurons via constrained nonlinear optimization” in Advances in Neural Information Processing Systems, NeurIPS 18 Conference Proceedings, eds. Y. Weiss, B. Scholkopf, J. Platt.
LinkOut - more resources
Full Text Sources

