Construction of a prognostic model and analysis of related mechanisms in breast cancer based on multiple datasets
- PMID: 40104717
- PMCID: PMC11912066
- DOI: 10.21037/tcr-24-838
Construction of a prognostic model and analysis of related mechanisms in breast cancer based on multiple datasets
Abstract
Background: Breast cancer (BC) is a common tumor among women and is a heterogeneous disease with many subtypes. Each subtype shows different clinical presentations, disease trajectories and prognoses, and different responses to neoadjuvant therapy; thus, a new and universal prognostic biomarker for BC patients is urgently needed. Our goal is to identify a novel prognostic molecular biomarker that can accurately predict the outcome of all BC subtypes and guide their clinical management.
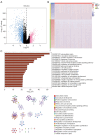
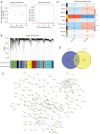
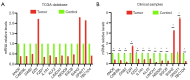
Methods: Utilizing data from The Cancer Genome Atlas (TCGA), we analyzed differential gene expression and patient clinical data. Weighted gene coexpression network analysis (WGCNA), Cox univariate regression and least absolute shrinkage and selection operator (LASSO) analysis were used to construct a prognostic model; the differential expression of the core genes in this model was validated via real-time quantitative polymerase chain reaction (RT-qPCR), and the reliability of the predictive model was validated in both an internal cohort and a BC patient dataset from the Gene Expression Omnibus (GEO) database. Further studies, such as gene set variation analysis (GSVA) and gene set enrichment analysis (GSEA), were performed to investigate the enrichment of signaling pathways. The CIBERSORT algorithm was used to estimate immune infiltration and tumor mutation burden (TMB), and drug sensitivity analysis was performed to evaluate the treatment response.
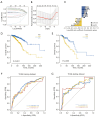
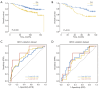
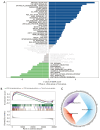
Results: A total of 1,643 differentially expressed genes were identified. After WGCNA and Cox regression combined with LASSO analysis, 15 genes were identified by screening and used to establish a prognostic gene signature. Further analysis revealed that the epithelial-mesenchymal transition (EMT) pathway gene signature was enriched in these genes. Each patient was assigned a risk score, and according to the median risk score, patients were classified into a high-risk group or a low-risk group. The prognosis of the low-risk group was better than that of the high-risk group (P<0.01), and analyses of two independent GEO validation cohorts yielded similar results. Furthermore, a nomogram was constructed and found to perform well in predicting prognosis. GSVA revealed that the EMT pathway, transforming growth factor β (TGF-β) signaling pathway and PI3K-Akt signaling pathway genes were enriched in the high-risk group, and the Wnt-β-catenin signaling pathway, DNA repair pathway and P53 pathway gene sets were enriched in the low-risk group. GSEA revealed genes related to TGF-β signaling and the PI3K-Akt signaling pathways were enriched in the high-risk group. CIBERSORT demonstrated that the low-risk group had greater infiltration of antitumor immune cells. The TMB and drug sensitivity results suggested that immunotherapy and chemotherapy are likely to be more effective in the low-risk group.
Conclusions: We established a new EMT pathway-related prognostic gene signature that can be used to effectively predict BC prognosis and treatment response.
Keywords: Breast cancer (BC); epithelial-mesenchymal transition pathway (EMT pathway); prognostic model.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-838/coif). The authors have no conflicts of interest to declare.
Figures



Similar articles
-
Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer.Front Immunol. 2022 Nov 21;13:1056932. doi: 10.3389/fimmu.2022.1056932. eCollection 2022. Front Immunol. 2022. PMID: 36479114 Free PMC article.
-
Comprehensive FGFR3 alteration-related transcriptomic characterization is involved in immune infiltration and correlated with prognosis and immunotherapy response of bladder cancer.Front Immunol. 2022 Jul 26;13:931906. doi: 10.3389/fimmu.2022.931906. eCollection 2022. Front Immunol. 2022. PMID: 35958598 Free PMC article.
-
Construction and validation of a bladder cancer risk model based on autophagy-related genes.Funct Integr Genomics. 2023 Jan 23;23(1):46. doi: 10.1007/s10142-022-00957-2. Funct Integr Genomics. 2023. PMID: 36689018
-
Identification of prognostic cancer-associated fibroblast markers in luminal breast cancer using weighted gene co-expression network analysis.Front Oncol. 2023 May 3;13:1191660. doi: 10.3389/fonc.2023.1191660. eCollection 2023. Front Oncol. 2023. PMID: 37207166 Free PMC article.
-
Development and Validation of a Novel Gene Signature for Predicting the Prognosis of Idiopathic Pulmonary Fibrosis Based on Three Epithelial-Mesenchymal Transition and Immune-Related Genes.Front Genet. 2022 Apr 26;13:865052. doi: 10.3389/fgene.2022.865052. eCollection 2022. Front Genet. 2022. PMID: 35559024 Free PMC article.
Cited by
-
Systematic Review and Meta-Analysis of AI-Assisted Mammography and the Systemic Immune-Inflammation Index in Breast Cancer: Diagnostic and Prognostic Perspectives.Medicina (Kaunas). 2025 Jun 27;61(7):1170. doi: 10.3390/medicina61071170. Medicina (Kaunas). 2025. PMID: 40731800 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
