A prognostic model for laryngeal squamous cell carcinoma based on the mitochondrial metabolism-related genes
- PMID: 40104737
- PMCID: PMC11912054
- DOI: 10.21037/tcr-24-1436
A prognostic model for laryngeal squamous cell carcinoma based on the mitochondrial metabolism-related genes
Abstract
Background: Mitochondrial metabolism-related genes (MMRGs) have emerged as potential therapeutic targets in cancer. This study aimed to construct a prognosis model based on MMRGs for patients with laryngeal squamous cell carcinoma (LSCC).
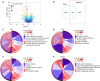
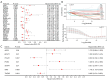
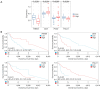
Methods: Differentially expressed MMRGs in LSCC were identified from The Cancer Genome Atlas (TCGA) and Molecular Signatures Database (MSigDB). Their functions were characterized by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). A prognostic model was established using univariate, least absolute shrinkage and selection operator (LASSO), and multivariate Cox regression analyses, and its performance was evaluated using Kaplan-Meier and receiver operating characteristic (ROC) curves. Gene set enrichment analysis (GSEA) was performed to elucidate the biological pathways associated with the hub prognostic MMRGs. Genetic perturbation similarity analysis (GPSA) was used to determine the regulatory network of hub genes. Additionally, the correlation of the hub MMRGs with the immune microenvironment and drug sensitivity was investigated.
Results: We identified 308 differentially expressed MMRGs, enriched in various metabolic processes and pathways. The prognostic model comprising four hub MMRGs (POLD1, PON2, SMS, and THEM5) accurately predicted patient outcomes, with the high-risk group exhibiting poorer survival. Additionally, high expression of POLD1 and THEM5 while low expression of PON2 and SMS indicated better prognosis for LSCC patients. GSEA revealed pathways correlated with each prognostic MMRG, such as PI3K-AKT-mTOR signaling pathways, while GPSA identified key regulatory genes interacting with four hub MMRGs. Furthermore, differences in the tumor immune microenvironment and somatic mutation profiles were observed between high- and low-risk groups. Finally, the correlation of four hub MMRGs with 30 drug sensitivity was revealed.
Conclusions: This study highlights the prognostic significance of MMRGs in LSCC and underscores their potential as biomarkers for LSCC therapy.
Keywords: Laryngeal squamous cell carcinoma (LSCC); immune microenvironment; mitochondrial metabolism-related gene (MMRG); mutation; prognosis.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1436/coif). The authors have no conflicts of interest to declare.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous
