Role of cancer stem cell heterogeneity in intrahepatic cholangiocarcinoma
- PMID: 40104739
- PMCID: PMC11912081
- DOI: 10.21037/tcr-24-1286
Role of cancer stem cell heterogeneity in intrahepatic cholangiocarcinoma
Abstract
Background: Intrahepatic cholangiocarcinoma (ICC) is a highly invasive bile duct cancer with poor prognosis due to frequent recurrence and limited effective treatments. Cancer stem cells (CSCs) contribute to ICC's therapeutic resistance and recurrence, driven by distinct cellular subpopulations with variable tumorigenic properties. Recent advances in single-cell RNA sequencing (scRNA-seq) have enabled a deeper exploration of cellular heterogeneity in tumors, offering insights into unique CSC subgroups that impact ICC progression and patient outcomes. This study aimed to investigate the effect of CSC heterogeneity on the prognosis of ICC.
Methods: The scRNA-seq dataset GSE142784 was retrieved from the Gene Expression Omnibus (GEO) database, and Bulk RNA-seq data were obtained from The Cancer Genome Atlas (TCGA) databases. Hallmarks and AUCell R package were adopted for analyzing the signaling pathway activity, CellChat for observing cell communication between subgroups, and SCENIC for analyzing transcription factors expression. The immune cell infiltration and drug sensitivity of the model were analyzed using the CIBERSORT algorithm and the "pRRophetic" R packages, respectively. And immunohistochemistry (IHC) tests were used to evaluate expression of transcription factors in ICC patients.
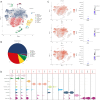
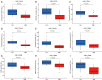
Results: Based on scRNA-seq data, five clusters (DLK+, CD13+, CD90+, CD133+, and other cholangiocarcinoma cells) were observed in ICC, which presented different signaling pathway activities, such as HSF1 and STAT1 were highly expressed in the CD133 cluster, and consistent with the results of IHC tests. Pathways like Notch and Wnt/β-catenin signaling transferred among above subgroups. Further, subgroups favored varied immune response and drug sensitivity, and CD133+ subgroup patients showed significantly shortened recurrence-free survival (RFS).
Conclusions: Configuring the subgroup of ICC is helpful for predicting the prognosis and drug resistance in ICC and can provide new strategies for cancer treatment.
Keywords: Intrahepatic cholangiocarcinoma (ICC); cancer stem cell heterogeneity (CSC heterogeneity); poor prognosis; therapeutic target.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1286/coif). The authors have no conflicts of interest to declare.
Figures





Similar articles
-
Clinical prognosticators and targets in the immune microenvironment of intrahepatic cholangiocarcinoma.Oncoimmunology. 2024 Oct 1;13(1):2406052. doi: 10.1080/2162402X.2024.2406052. eCollection 2024. Oncoimmunology. 2024. PMID: 39359389 Free PMC article.
-
LGR5 induces β-catenin activation and augments tumour progression by activating STAT3 in human intrahepatic cholangiocarcinoma.Liver Int. 2021 Apr;41(4):865-881. doi: 10.1111/liv.14747. Epub 2020 Dec 9. Liver Int. 2021. PMID: 33249719
-
YBX1 promotes stemness and cisplatin insensitivity in intrahepatic cholangiocarcinoma via the AKT/β-catenin axis.J Gene Med. 2024 May;26(5):e3689. doi: 10.1002/jgm.3689. J Gene Med. 2024. PMID: 38676365
-
The role of Tripartite motif containing 59 (TRIM59) in the proliferation and prognosis of intrahepatic cholangiocarcinoma.Pathol Res Pract. 2022 Aug;236:153989. doi: 10.1016/j.prp.2022.153989. Epub 2022 Jun 17. Pathol Res Pract. 2022. PMID: 35753134 Review.
-
Multiple cellular origins and molecular evolution of intrahepatic cholangiocarcinoma.Cancer Lett. 2016 Sep 1;379(2):253-61. doi: 10.1016/j.canlet.2016.02.038. Epub 2016 Mar 2. Cancer Lett. 2016. PMID: 26940139 Review.
References
-
- Lee AJ, Chun YS. Intrahepatic cholangiocarcinoma: the AJCC/UICC 8th edition updates. Chin Clin Oncol 2018;7:52. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
