Comprehensive bioinformatics analysis of co-mutation of FLG2 and TP53 reveals prognostic effect and influences on the immune infiltration in ovarian serous cystadenocarcinoma
- PMID: 40104745
- PMCID: PMC11912057
- DOI: 10.21037/tcr-24-1596
Comprehensive bioinformatics analysis of co-mutation of FLG2 and TP53 reveals prognostic effect and influences on the immune infiltration in ovarian serous cystadenocarcinoma
Abstract
Background: Ovarian cancer remains one of the most lethal gynecological malignancies, characterized by late-stage diagnosis and high rates of recurrence. The present study aims to explore the prognostic and immunological implications of FLG2 and TP53, the two genes exhibiting a high mutation frequency across various cancer types, in the context of ovarian serous cystadenocarcinoma (OV).
Methods: The study systematically analyzed and discussed the potential implications of co-mutation of FLG2 and TP53 on prognosis and immune response using a cohort of 585 ovarian cancer samples. The differentially expressed genes (DEGs) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed on 300 ovarian cancer samples with RNA sequencing (RNA-seq) data.
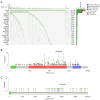
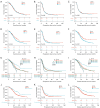
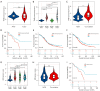
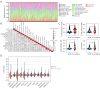
Results: The co-mutation of FLG2 and TP53 was identified in the 585 ovarian cancer cohort, and the group with co-mutation exhibited improved outcomes in terms of overall survival (OS), progression-free survival (PFS), and disease-specific survival (DSS). Additionally, the co-mutation (FLG2 +/TP53 +) group demonstrated higher scores in tumor mutation burden (TMB) comparing to that of the other three groups. The score of microsatellite instability (MSI) in the co-mutant group was only higher than that of the co-wild-type (FLG2 -/TP53 -). A total of 327 DEGs were identified in both the co-mutation and non-co-mutation (NCM) groups using limma analysis in the subgroup of 300 patients with RNA-seq data. Subsequent KEGG analysis revealed that these DEGs were implicated in various biological processes, including thermogenesis, Parkinson's disease (PD), and oxidative phosphorylation signaling pathways. Additionally, the co-mutation group exhibited elevated levels of various immune cells. Furthermore, a nomogram with high predictive accuracy was developed by integrating co-mutation status with clinical characteristics.
Conclusions: In the context of OV, the concurrent mutation of FLG2 and TP53 not only induces immune activation, but also helps identify a subset of patients with a more favorable prognosis.
Keywords: FLG2/TP53; Ovarian serous cystadenocarcinoma (OV); co-mutation; immune infiltration; prognosis.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1596/coif). The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Exploration of Key Genes Combining with Immune Infiltration Level andTumor Mutational Burden in Hepatocellular Carcinoma.Comb Chem High Throughput Screen. 2024;27(14):2110-2124. doi: 10.2174/0113862073239916231023053142. Comb Chem High Throughput Screen. 2024. PMID: 38213141
-
Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers.Comput Struct Biotechnol J. 2022 Jun 18;20:3106-3119. doi: 10.1016/j.csbj.2022.06.039. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35782736 Free PMC article.
-
ANXA2P2: A Potential Immunological and Prognostic Signature in Ovarian Serous Cystadenocarcinoma via Pan-Carcinoma Synthesis.Front Oncol. 2022 Feb 8;12:818977. doi: 10.3389/fonc.2022.818977. eCollection 2022. Front Oncol. 2022. PMID: 35211410 Free PMC article.
-
A special prognostic indicator: tumor mutation burden combined with immune infiltrates in lung adenocarcinoma with TP53 mutation.Transl Cancer Res. 2021 Sep;10(9):3963-3978. doi: 10.21037/tcr-21-565. Transl Cancer Res. 2021. PMID: 35116695 Free PMC article.
-
Identifies microtubule-binding protein CSPP1 as a novel cancer biomarker associated with ferroptosis and tumor microenvironment.Comput Struct Biotechnol J. 2022 Jun 24;20:3322-3335. doi: 10.1016/j.csbj.2022.06.046. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832625 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
