The N6-methyladenosine reader IGF2BP3 promotes bladder cancer progression through enhancing HSP90AB1 expression
- PMID: 40105114
- PMCID: PMC12220844
- DOI: 10.1111/febs.70068
The N6-methyladenosine reader IGF2BP3 promotes bladder cancer progression through enhancing HSP90AB1 expression
Abstract
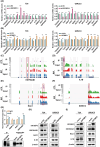
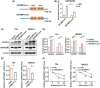
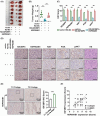
N6-methyladenosine (m6A) is the most abundant RNA modification in mammalian cells, and has emerged as an important player in tumour development through post-transcriptional gene regulation. In this study, we found that the m6A reader protein IGF2BP3 was the most upregulated m6A modifier in bladder cancer through the proteomic analysis of 17 pairs of human bladder cancer tissues and adjacent normal bladder tissues, for which the expression was also positively correlated with higher tumour stage and poorer prognosis. In vitro and in vivo assays demonstrated the powerful oncogenic function of IGF2BP3 in bladder cancer. Further combined analyses of RNA-sequencing, m6A-sequencing, and RIP (RNA Binding Protein Immunoprecipitation)-sequencing, as well as site-directed mutagenesis assays and RIP-qPCR identified m6A-tagged HSP90AB1 mRNA as a direct target of IGF2BP3. Mechanistically, through in vitro and in vivo assays, as well as clinical sample analysis, we demonstrated that IGF2BP3 modulated the expression of HSP90AB1 in an m6A modification-dependent manner, thus activating the PI3K/AKT-signaling pathway, and promoting the development of bladder cancer. Collectively, our study highlights the critical role of the IGF2BP3-HSP90AB1-signaling axis in bladder cancer progression, which may serve as a promising therapeutic approach for bladder cancer.
Keywords: HSP90AB1; IGF2BP3; PI3K/AKT signaling; bladder cancer; m6A.
© 2025 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Multi-omics analysis unveils the predictive value of IGF2BP3/SPHK1 signaling in cancer stem cells for prognosis and immunotherapeutic response in muscle-invasive bladder cancer.J Transl Med. 2024 Oct 4;22(1):900. doi: 10.1186/s12967-024-05685-8. J Transl Med. 2024. PMID: 39367493 Free PMC article.
-
m6A-YTHDF1 Mediated Regulation of GRIN2D in Bladder Cancer Progression and Aerobic Glycolysis.Biochem Genet. 2025 Aug;63(4):3220-3239. doi: 10.1007/s10528-024-10875-6. Epub 2024 Jun 29. Biochem Genet. 2025. PMID: 38951355
-
Hsa_circ_0005320 affects cell proliferation and the cell cycle via the IGF2BP3/CDK2 axis in bladder cancer.Cell Signal. 2024 Jul;119:111154. doi: 10.1016/j.cellsig.2024.111154. Epub 2024 Mar 31. Cell Signal. 2024. PMID: 38565412
-
Dysregulated PI3K/AKT signaling in oral squamous cell carcinoma: The tumor microenvironment and epigenetic modifiers as key drivers.Oncol Res. 2025 Jul 18;33(8):1835-1860. doi: 10.32604/or.2025.064010. eCollection 2025. Oncol Res. 2025. PMID: 40746882 Free PMC article. Review.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A & Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71, 209–249. - PubMed
-
- Gore JL & Wright JL (2019) Can we prevent bladder cancer recurrences? Eur Urol 75, 602–603. - PubMed
-
- Knowles MA & Hurst CD (2015) Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 15, 25–41. - PubMed
-
- Patel VG, Oh WK & Galsky MD (2020) Treatment of muscle‐invasive and advanced bladder cancer in 2020. CA Cancer J Clin 70, 404–423. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

