Discovery of circular transcripts of the human BCL2-like 12 (BCL2L12) apoptosis-related gene, using targeted nanopore sequencing, provides new insights into circular RNA biology
- PMID: 40106061
- PMCID: PMC11923030
- DOI: 10.1007/s10142-025-01578-1
Discovery of circular transcripts of the human BCL2-like 12 (BCL2L12) apoptosis-related gene, using targeted nanopore sequencing, provides new insights into circular RNA biology
Abstract
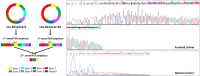
Circular RNAs (circRNAs) constitute an RNA type formed by back-splicing. BCL2-like 12 (BCL2L12) is an apoptosis-related gene comprising 7 exons. In this study, we used targeted nanopore sequencing to identify circular BCL2L12 transcripts in human colorectal cancer cells and investigated the effect of circRNA silencing on mRNA expression of the parental gene. In brief, nanopore sequencing following nested PCR amplification of cDNAs of BCL2L12 circRNAs from 7 colorectal cancer cell lines unraveled 46 BCL2L12 circRNAs, most of which described for the first time. Interestingly, 40 novel circRNAs are likely to form via back-splicing between non-canonical back-splice sites residing in highly similar regions of the primary transcripts. All back-splice junctions were validated using next-generation sequencing (NGS) after circRNA enrichment. Surprisingly, 2 novel circRNAs also comprised a poly(A) tract after BCL2L12 exon 7; this poly(A) tract was back-spliced to exon 1, in both cases. Furthermore, the selective silencing of a BCL2L12 circRNA resulted in a subsequent decrease of BCL2L12 mRNA levels in HCT 116 cells, thus providing evidence of parental gene expression regulation by circRNAs. In conclusion, our study led to the discovery of many circular transcripts from a single human gene and provided new insights into circRNA biogenesis and mode of action.
Keywords: Alternative back-splicing; Alternative splicing; CircRNAs; Colorectal cancer; Gene expression regulation; Third-generation (long-read) sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: Prof. Dr. Christos K. Kontos is an Editor for Functional and Integrative Genomics. The authors have no relevant financial or non-financial interests to disclose.
Figures





References
-
- Adamopoulos PG, Kontos CK, Tsiakanikas P, Scorilas A (2016) Identification of novel alternative splice variants of the BCL2L12 gene in human cancer cells using next-generation sequencing methodology. Cancer Lett 373(1):119–129. 10.1016/j.canlet.2016.01.019 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

