Structural basis for the substrate recognition and transport mechanism of the human y+LAT1-4F2hc transporter complex
- PMID: 40106545
- PMCID: PMC11922002
- DOI: 10.1126/sciadv.adq0558
Structural basis for the substrate recognition and transport mechanism of the human y+LAT1-4F2hc transporter complex
Abstract
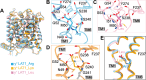
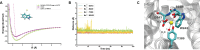
Heteromeric amino acid transporters (HATs), including y+LAT1-4F2hc complex, are responsible for transporting amino acids across membranes, and mutations in y+LAT1 cause lysinuric protein intolerance (LPI), a hereditary disorder characterized by defective cationic amino acid transport. The relationship between LPI and specific mutations in y+LAT1 has yet to be fully understood. In this study, we characterized the function of y+LAT1-4F2hc complex in mammalian cells and determined the cryo-EM structures of the human y+LAT1-4F2hc complex in two distinct conformations: the apo state in an inward-open conformation and the native substrate-bound state in an outward-open conformation. Structural analysis suggests that Asp243 in y+LAT1 plays a crucial role in coordination with sodium ion and substrate selectivity. Molecular dynamic (MD) simulations further revealed the different transport mechanism of cationic amino acids and neutral amino acids. These results provide important insights into the mechanisms of the substrate binding and working cycle of HATs.
Figures





References
-
- Bröer S., Palacin M., The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 436, 193–211 (2011). - PubMed
-
- Kandasamy P., Gyimesi G., Kanai Y., Hediger M. A., Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 43, 752–789 (2018). - PubMed
-
- Fotiadis D., Kanai Y., Palacin M., The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34, 139–158 (2013). - PubMed
-
- Mastroberardino L., Spindler B., Pfeiffer R., Skelly P. J., Loffing J., Shoemaker C. B., Verrey F., Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature 395, 288–291 (1998). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

