In a nutshell: pistachio genome and kernel development
- PMID: 40107319
- PMCID: PMC11982797
- DOI: 10.1111/nph.70060
In a nutshell: pistachio genome and kernel development
Abstract
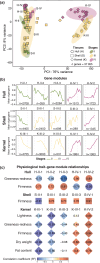
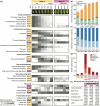
Pistachio is a sustainable nut crop with exceptional climate resilience and nutritional value. However, the molecular processes underlying pistachio nut development and nutritional traits are largely unknown, compounded by limited genomic and molecular resources. To advance pistachios as a future food source and a model system for hard-shelled fruits, we generated a chromosome-scale reference genome of the most widely grown pistachio cultivar (Pistacia vera 'Kerman') and a spatiotemporal study of nut development. We integrated tissue-level physiological data from thousands of nuts over three growing seasons with transcriptomic data encompassing 14 developmental time points of the hull, shell, and kernel to assemble gene modules associated with physiological changes. Our study defined four distinct stages of pistachio nut growth and maturation. We then focused on the kernel to identify transcriptional and metabolic changes in molecular pathways governing nutritional quality, such as the accumulation of unsaturated fatty acids, which are vital for shelf life and dietary value. These findings revealed key candidate conserved regulatory genes, such as PvAP2-WRI1 and PvNFYB-LEC1, likely involved in oil accumulation in kernels. This work yields new knowledge and resources that will inform other woody crops and facilitate further improvement of pistachio as a globally significant, sustainable, and nutritious crop.
Keywords: Kerman; Pistacia vera; chromosome‐scale assembly; kernel metabolic profile; nut development; nut physiology; pistachio; reference genome; spatiotemporal transcriptome; tree crop.
© 2025 The Author(s). New Phytologist © 2025 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures



References
-
- Andrews S. 2010. fastqc: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed‐effects models Usinglme4. Journal of Statistical Software 67: 1–48.
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B, Statistical Methodology 57: 289–300.

