An alternative adaptation strategy of the CCA-adding enzyme to accept noncanonical tRNA substrates in Ascaris suum
- PMID: 40107618
- PMCID: PMC12013499
- DOI: 10.1016/j.jbc.2025.108414
An alternative adaptation strategy of the CCA-adding enzyme to accept noncanonical tRNA substrates in Ascaris suum
Abstract
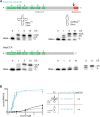
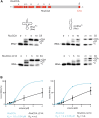
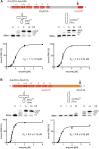
Playing a central role in translation, tRNAs act as an essential adapter linking the correct amino acid to the corresponding mRNA codon in translation. Due to this function, all tRNAs exhibit a typical secondary and tertiary structure to be recognized by the tRNA maturation enzymes as well as many components of the translation machinery. Yet, there is growing evidence for structurally deviating tRNAs in metazoan mitochondria, requiring a coevolution and adaptation of these enzymes to the unusual structures of their substrates. Here, it is shown that the CCA-adding enzyme of Ascaris suum carries such a specific adaptation in form of a C-terminal extension. The corresponding enzymes of other nematodes also carry such extensions, and many of them have an additional adaptation in a small region of their N-terminal catalytic core. Thus, the presented data indicate that these enzymes evolved two distinct strategies to tolerate noncanonical tRNAs as substrates for CCA incorporation. The identified C-terminal extension represents a surprising case of convergent evolution in tRNA substrate adaptation, as the nematode mitochondrial translation factor EF-Tu1 carries a similar extension that is essential for efficient binding to such structurally deviating tRNAs.
Keywords: CCA-adding enzyme; coevolution; convergent evolution; miniaturized tRNAs; noncanonical tRNAs; tRNA nucleotidyltransferase.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Giegé R., Jühling F., Pütz J., Stadler P., Sauter C., Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Inter. Rev. RNA. 2012;3:37–61. - PubMed
-
- Kim S.H., Suddath F.L., Quigley G.J., McPherson A., Sussmann J.L., Wang A.H., et al. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. - PubMed
-
- Sprinzl M., Cramer F. The -C-C-A end of tRNA and its role in protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1979;22:1–69. - PubMed
-
- Green R., Noller H.F. Ribosomes and translation. Annu. Rev. Biochem. 1997;66:679–716. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

