Dipeptidyl peptidase DPF-3 is a gatekeeper of microRNA Argonaute compensation in animals
- PMID: 40108168
- PMCID: PMC11923051
- DOI: 10.1038/s41467-025-58141-6
Dipeptidyl peptidase DPF-3 is a gatekeeper of microRNA Argonaute compensation in animals
Abstract
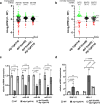
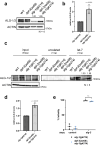
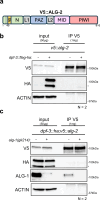
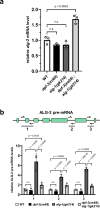
MicroRNAs (miRNAs) are essential regulators involved in multiple biological processes. To achieve their gene repression function, they are loaded in miRNA-specific Argonautes to form the miRNA-induced silencing complex (miRISC). Mammals and C. elegans possess more than one paralog of miRNA-specific Argonautes, but the dynamic between them remains unclear. Here, we report the conserved dipeptidyl peptidase DPF-3 as an interactor of the miRNA-specific Argonaute ALG-1 in C. elegans. Knockout of dpf-3 increases ALG-2 levels and miRISC formation in alg-1 loss-of-function animals, thereby compensating for ALG-1 loss and rescuing miRNA-related defects observed. DPF-3 can cleave an ALG-2 N-terminal peptide in vitro but does not appear to rely on this catalytic activity to regulate ALG-2 in vivo. This study uncovers the importance of DPF-3 in the miRNA pathway and provides insights into how multiple miRNA Argonautes contribute to achieving proper miRNA-mediated gene regulation in animals.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Eulalio, A., Huntzinger, E. & Izaurralde, E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol.15, 346–353 (2008). - PubMed
-
- Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell116, 281–297 (2004). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

