Clade-specific extracellular vesicles from Akkermansia muciniphila mediate competitive colonization via direct inhibition and immune stimulation
- PMID: 40108178
- PMCID: PMC11923206
- DOI: 10.1038/s41467-025-57631-x
Clade-specific extracellular vesicles from Akkermansia muciniphila mediate competitive colonization via direct inhibition and immune stimulation
Abstract
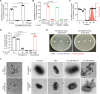
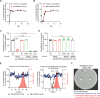
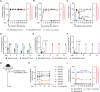
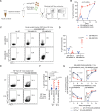
Akkermansia muciniphila, a promising candidate for next-generation probiotics, exhibits significant genomic diversity, classified into several distinct clades (AmI to AmIV). Notably, a single Akkermansia clade tends to predominate within individual hosts, with co-occurrence of different clades being rare. The mechanisms driving such clade-specific exclusion remain unclear. Here, we show that extracellular vesicles (EVs) derived from AmII clade inhibit the growth of clade I (AmI), conferring a competitive advantage to AmII. Moreover, we observe clade-specific immunoglobulin A (IgA) responses, where AmII clade-specific IgAs, induced by EVs from AmII, facilitate niche occupancy and competitive exclusion of AmI. These findings provide insights into the competitive dynamics of A. muciniphila clades and suggest that future personalized microbiome interventions could be optimized by considering the clade composition of A. muciniphila in individual hosts.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M-G.H., D.H.K., D.K.L., S-N.L., and J-G.S. were employed by Enterobiome Inc. The authors declare that this study did not receive funding from Enterobiome Inc. The remaining authors declare no competing interests.
Figures


References
-
- Wang, B., Yao, M., Lv, L., Ling, Z. & Li, L. The human microbiota in health and disease. Engineering3, 71–82 (2017). - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

