Histological signatures map anti-fibrotic factors in mouse and human lungs
- PMID: 40108456
- PMCID: PMC12105817
- DOI: 10.1038/s41586-025-08727-3
Histological signatures map anti-fibrotic factors in mouse and human lungs
Abstract
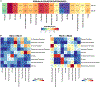
Fibrosis, the replacement of healthy tissue with collagen-rich matrix, can occur following injury in almost every organ1,2. Mouse lungs follow a stereotyped sequence of fibrogenesis-to-resolution after bleomycin injury3, and we reasoned that profiling post-injury histological stages could uncover pro-fibrotic versus anti-fibrotic features with functional value for human fibrosis. Here we quantified spatiotemporally resolved matrix transformations for integration with multi-omic data. First, we charted stepwise trajectories of matrix aberration versus resolution, derived from a high-dimensional set of histological fibre features, that denoted a reversible transition in uniform-to-disordered histological architecture. Single-cell sequencing along these trajectories identified temporally enriched 'ECM-secreting' (Csmd1-expressing) and 'pro-resolving' (Cd248-expressing) fibroblasts at the respective post-injury stages. Visium-based spatial analysis further suggested divergent matrix architectures and spatial-transcriptional neighbourhoods by fibroblast subtype, identifying distinct fibrotic versus non-fibrotic biomolecular milieu. Critically, pro-resolving fibroblast instillation helped to ameliorate fibrosis in vivo. Furthermore, the fibroblast neighbourhood-associated factors SERPINE2 and PI16 functionally modulated human lung fibrosis ex vivo. Spatial phenotyping of idiopathic pulmonary fibrosis at protein level additionally uncovered analogous fibroblast subtypes and neighbourhoods in human disease. Collectively, these findings establish an atlas of pro- and anti-fibrotic factors that underlie lung matrix architecture and implicate fibroblast-associated biological features in modulating fibrotic progression versus resolution.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



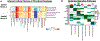

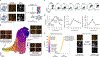





References
-
- Glassberg MK Overview of idiopathic pulmonary fibrosis, evidence-based guidelines, and recent developments in the treatment landscape. Am. J. Manag. Care 25, S195–S203 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

