Kit-mediated autophagy suppression driven by a viral oncoprotein emerges as a crucial survival mechanism in Merkel cell carcinoma
- PMID: 40108758
- PMCID: PMC12283026
- DOI: 10.1080/15548627.2025.2477385
Kit-mediated autophagy suppression driven by a viral oncoprotein emerges as a crucial survival mechanism in Merkel cell carcinoma
Abstract
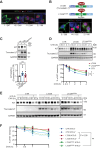
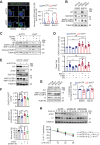
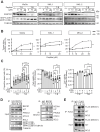
The KIT/c-KIT proto-oncogene is frequently over-expressed in Merkel cell carcinoma (MCC), an aggressive skin cancer commonly caused by Merkel cell polyomavirus (MCPyV). Here, we demonstrated that truncated MCPyV-encoded large T-antigen (LT) suppressed macroautophagy/autophagy by stabilizing and sequestering KIT in the paranuclear compartment via binding VPS39. KIT engaged with phosphorylated BECN1, thereby enhancing its association with BCL2 while diminishing its interaction with the PIK3C3 complex. This process ultimately resulted in the suppression of autophagy. Depletion of KIT triggered both autophagy and apoptosis, and decreased LT expression. Conversely, blocking autophagy in KIT-depleted cells restored LT levels and rescued apoptosis. Additionally, stimulating autophagy efficiently increased cell death and inhibited tumor growth of MCC xenografts in mice. These insights into the interplay between MCPyV LT and autophagy regulation reveal important mechanisms by which viral oncoproteins are essential for MCC cell viability. Thus, autophagy-inducing agents represent a therapeutic strategy in advanced MCPyV-associated MCC.Abbreviation: 3-MA, 3-methyladenine; AL, autolysosome; AP, autophagosome; Baf-A1, bafilomycin A1; BARA, β-α repeated autophagy specific domain; BH3, BCL2 homology 3 domain; CCD, coiled-coil domain; CHX, cycloheximide; Co-IP, co-immunoprecipitation; CQ, chloroquine; CTR, control; DAPI, 4',6-diamidino-2-phenylindole; EBSS, Earle's balanced salt solution; ECD, evolutionarily conserved domain; EEE, three-tyrosine phosphomimetic mutations Y229E Y233E Y352E; ER, endoplasmic reticulum; FFF, three-tyrosine non-phosphomimetic mutations; FFPE, formalin-fixed paraffin-embedded; FL, full-length; GIST, gastrointestinal stromal tumor; IB, immunoblotting; IHC, immunohistochemistry; KIT-HEK293, KIT stably expressing HEK293 cells; KRT20/CK20, keratin 20; LT, large T-antigen; LT339, MCPyV truncated LT antigen; LTco, codon-optimized MCPyV LT antigen; MCC, Merkel cell carcinoma; MCPyV-, MCPyV-negative; MCPyV, Merkel cell polyomavirus; MCPyV+, MCPyV-positive; PARP1, poly(ADP-ribose) polymerase 1; PCI, pan-caspase inhibitor; PI, propidium iodide; PtdIns3K, class III phosphatidylinositol 3-kinase; PtdIns3P, phosphatidylinositol-3-phosphate; RB1, RB transcriptional corepressor 1; RTKs, receptor tyrosine kinases; KITLG/SCF, KIT ligand; sT, small T-antigen; sTco, codon-optimized MCPyV sT antigen; T-B, Tat-BECN1; T-S, Tat-scrambled; TEM, transmission electron microscopy.
Keywords: Autophagy; BECN1; KIT; Merkel cell carcinoma; Merkel cell polyomavirus; large T antigen.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
