Genomic and Transcriptomic Characterization of Protein Kinase C Fusion Melanocytic Neoplasms With Distinctive Hypopigmented Histomorphology: A Single-Institution Study
- PMID: 40109002
- PMCID: PMC12061626
- DOI: 10.1111/cup.14801
Genomic and Transcriptomic Characterization of Protein Kinase C Fusion Melanocytic Neoplasms With Distinctive Hypopigmented Histomorphology: A Single-Institution Study
Abstract
Background: Genomic fusions involving Protein Kinase C (PKC or PRKC) have been classically identified in a subset of melanocytic neoplasms with heavy melanin pigmentation as described in older series. They were recently reclassified from the pigmented epithelioid melanocytoma (PEM) category to the blue nevus (BN) category in the fifth edition of the World Health Organization (WHO) Classification of Skin Tumors.
Methods: Herein, we report a series of eight mostly hypopigmented PRKC fusion melanocytic tumors with novel comprehensive molecular characterization. Clinical, histopathologic, and immunohistochemical findings were reviewed. Next-generation sequencing (NGS) data on genomic and transcriptomic levels were explored.
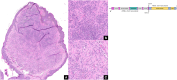
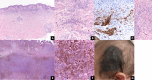
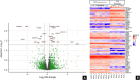
Results: Histomorphology showed a biphasic pattern with hypercellular areas and hypocellular areas with dense fibrotic stroma and collagen trapping. The clinical courses were uncomplicated after excisions. NGS revealed three cases of PRKCB fusion and five cases of PRKCA fusions. RNA differential analysis against six blue nevi showed a group of genes with significantly higher transcription levels and strong enrichment in the direct p53 effectors gene set. PRKC fusion tumors also demonstrated significantly stronger p53 IHC staining.
Conclusion: We further expand the morphologic spectrum of PRKC fusion melanocytic tumors and provide insight into their morphologic identification. Our novel transcriptome-level findings provide insight into the nuanced molecular events and new evidence for classification.
Keywords: Protein Kinase C; melanocytic neoplasm; molecular pathology.
© 2025 The Author(s). Journal of Cutaneous Pathology published by John Wiley & Sons Ltd.
Conflict of interest statement
Brandon Umphress is a consultant pathologist for Tempus Labs, however, unrelated to this work. No other conflicts to disclose.
Figures



References
-
- Elder D. E., Bastian B. C., Cree I. A., Massi D., and Scolyer R. A., “The 2018 World Health Organization Classification of Cutaneous, Mucosal, and Uveal Melanoma: Detailed Analysis of 9 Distinct Subtypes Defined by Their Evolutionary Pathway,” Archives of Pathology & Laboratory Medicine 144, no. 4 (2020): 500–522, 10.5858/arpa.2019-0561-RA. - DOI - PubMed
-
- Cohen J. N., Joseph N. M., North J. P., Onodera C., Zembowicz A., and LeBoit P. E., “Genomic Analysis of Pigmented Epithelioid Melanocytomas Reveals Recurrent Alterations in PRKAR1A, and PRKCA Genes,” American Journal of Surgical Pathology 41, no. 10 (2017): 1333–1346, 10.1097/PAS.0000000000000902. - DOI - PubMed
-
- Zembowicz A., Carney J. A., and Mihm M. C., “Pigmented Epithelioid Melanocytoma: A Low‐Grade Melanocytic Tumor With Metastatic Potential Indistinguishable From Animal‐Type Melanoma and Epithelioid Blue Nevus,” American Journal of Surgical Pathology 28, no. 1 (2004): 31–40, 10.1097/00000478-200401000-00002. - DOI - PubMed
-
- Zembowicz A., Knoepp S. M., Bei T., et al., “Loss of Expression of Protein Kinase a Regulatory Subunit 1alpha in Pigmented Epithelioid Melanocytoma But Not in Melanoma or Other Melanocytic Lesions,” American Journal of Surgical Pathology 31, no. 11 (2007): 1764–1775, 10.1097/PAS.0b013e318057faa7. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

