Integrating sequencing methods with machine learning for antimicrobial susceptibility testing in pediatric infections: current advances and future insights
- PMID: 40109965
- PMCID: PMC11919855
- DOI: 10.3389/fmicb.2025.1528696
Integrating sequencing methods with machine learning for antimicrobial susceptibility testing in pediatric infections: current advances and future insights
Abstract
Antimicrobial resistance (AMR) presents a critical challenge in clinical settings, particularly among pediatric patients with life-threatening conditions such as sepsis, meningitis, and neonatal infections. The increasing prevalence of multi- and pan-resistant pathogens is strongly associated with adverse clinical outcomes. Recent technological advances in sequencing methods, including metagenomic next-generation sequencing (mNGS), Oxford Nanopore Technologies (ONT), and targeted sequencing (TS), have significantly enhanced the detection of both pathogens and their associated resistance genes. However, discrepancies between resistance gene detection and antimicrobial susceptibility testing (AST) often hinder the direct clinical application of sequencing results. These inconsistencies may arise from factors such as genetic mutations or variants in resistance genes, differences in the phenotypic expression of resistance, and the influence of environmental conditions on resistance levels, which can lead to variations in the observed resistance patterns. Machine learning (ML) provides a promising solution by integrating large-scale resistance data with sequencing outcomes, enabling more accurate predictions of pathogen drug susceptibility. This review explores the application of sequencing technologies and ML in the context of pediatric infections, with a focus on their potential to track the evolution of resistance genes and predict antibiotic susceptibility. The goal of this review is to promote the incorporation of ML-based predictions into clinical practice, thereby improving the management of AMR in pediatric populations.
Keywords: Oxford Nanopore Technologies; antimicrobial resistance; machine learning; next-generation sequencing; pediatrics.
Copyright © 2025 Zou, Tang, Qiao, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
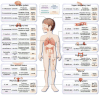


Similar articles
-
Automated antimicrobial susceptibility testing and antimicrobial resistance genotyping using Illumina and Oxford Nanopore Technologies sequencing data among Enterobacteriaceae.Front Microbiol. 2022 Aug 8;13:973605. doi: 10.3389/fmicb.2022.973605. eCollection 2022. Front Microbiol. 2022. PMID: 36003946 Free PMC article.
-
Metagenomic prediction of antimicrobial resistance in critically ill patients with lower respiratory tract infections.Genome Med. 2022 Jul 12;14(1):74. doi: 10.1186/s13073-022-01072-4. Genome Med. 2022. PMID: 35818068 Free PMC article.
-
Large-scale genomic analysis reveals significant role of insertion sequences in antimicrobial resistance of Acinetobacter baumannii.mBio. 2025 Mar 12;16(3):e0285224. doi: 10.1128/mbio.02852-24. Epub 2025 Feb 20. mBio. 2025. PMID: 39976435 Free PMC article.
-
Machine Learning Approaches for Microorganism Identification, Virulence Assessment, and Antimicrobial Susceptibility Evaluation Using DNA Sequencing Methods: A Systematic Review.Mol Biotechnol. 2024 Nov 9. doi: 10.1007/s12033-024-01309-0. Online ahead of print. Mol Biotechnol. 2024. PMID: 39520638 Review.
-
Advancements in AI-driven drug sensitivity testing research.Front Cell Infect Microbiol. 2025 May 2;15:1560569. doi: 10.3389/fcimb.2025.1560569. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40384974 Free PMC article. Review.
References
-
- Alygizakis N., Ng K., Čirka Ľ, Berendonk T., Cerqueira F., Cytryn E., et al. (2024). Making waves: The NORMAN antibiotic resistant bacteria and resistance genes database (NORMAN ARB&ARG)–An invitation for collaboration to tackle antibiotic resistance. Water Res. 257:121689. 10.1016/j.watres.2024.121689 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

