Comparative analysis of the microbial composition of three packaged sliced dry-cured hams from a Chinese market
- PMID: 40109974
- PMCID: PMC11919833
- DOI: 10.3389/fmicb.2025.1531005
Comparative analysis of the microbial composition of three packaged sliced dry-cured hams from a Chinese market
Abstract
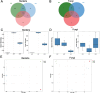
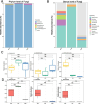
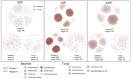
Ham, a widely consumed and culturally significant food, undergoes fermentation and aging processes that contribute to its distinctive flavor and texture. These processes are influenced by a complex interplay of microbial communities, which vary by the production region. Understanding these microbial dynamics can provide insights into flavor development and quality improvements in ham. In this study, the microbial communities found in ham produced in three distinct regions were compared, revealing that bacteria have a more dominant role in shaping the overall microbiota than fungi. Notably, each type of ham exhibited a unique microbial profile, although those from similar regions shared more similar profiles. Specific bacterial biomarkers were identified for each regional ham: Lactobacillus and Tetragonococcus in Serrano prosciutto, Odoribacter, Alistipes, Staphylococcus, and Akkermansia in Jinhua prosciutto, and Pseudomonas, Blautia, and Bacteroides in Xuanwei prosciutto. The microbial network analysis identified closer associations between microorganisms in the domestically produced Chinese hams than in the Spanish ham, suggesting limited foreign microbial invasions that contributed to a richer, more stable flavor. These findings offer new insights into how microbial interactions shape the development of flavor and quality in ham and clarify future strategies for improving the production process by leveraging microbial communities.
Keywords: Jinhua prosciutto; Serrano prosciutto; Xuanwei prosciutto; microbial composition; packaged sliced dry-cured hams.
Copyright © 2025 Luo and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Effect of high-pressure treatment on the survival of Listeria monocytogenes Scott A in sliced vacuum-packaged Iberian and Serrano cured hams.J Food Prot. 2006 Oct;69(10):2539-43. doi: 10.4315/0362-028x-69.10.2539. J Food Prot. 2006. PMID: 17066942
-
Study on the Changes and Correlation of Microorganisms and Flavor in Different Processing Stages of Mianning Ham.Foods. 2024 Aug 18;13(16):2587. doi: 10.3390/foods13162587. Foods. 2024. PMID: 39200514 Free PMC article.
-
Role of microbiota and its ecological succession on flavor formation in traditional dry-cured ham: a review.Crit Rev Food Sci Nutr. 2025;65(5):992-1008. doi: 10.1080/10408398.2023.2286634. Epub 2023 Dec 9. Crit Rev Food Sci Nutr. 2025. PMID: 38069684 Review.
-
Changes in the extent and products of In vitro protein digestion during the ripening periods of Chinese dry-cured hams.Meat Sci. 2021 Jan;171:108290. doi: 10.1016/j.meatsci.2020.108290. Epub 2020 Aug 26. Meat Sci. 2021. PMID: 32949821
-
Factors in pig production that impact the quality of dry-cured ham: a review.Animal. 2012 Feb;6(2):327-38. doi: 10.1017/S1751731111001625. Animal. 2012. PMID: 22436192 Review.
References
-
- Bastian M., Heymann S., Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 3, 361–362. doi: 10.1609/icwsm.v3i1.13937 - DOI
LinkOut - more resources
Full Text Sources

