Renal parenchymal volume analysis: Clinical and research applications
- PMID: 40109982
- PMCID: PMC11922601
- DOI: 10.1002/bco2.70013
Renal parenchymal volume analysis: Clinical and research applications
Abstract
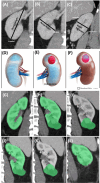
Background and objectives: In most patients, the renal parenchymal volumes in each kidney directly correlate with function and can be used as a proxy to determine split renal function (SRF). This simple principle forms the basis for parenchymal volume analysis (PVA) with semiautomated software, which can be leveraged to predict SRF and new-baseline glomerular filtration rate (NBGFR) following nephrectomy. PVA was originally used to evaluate renal transplantation donors and has replaced nuclear renal scans (NRS) in this domain. PVA has subsequently been explored for the management of patients with kidney cancer for whom difficult decisions about radical versus partial nephrectomy can be influenced by accurate prediction of NBGFR. Our objective is to present a comprehensive review of the applications of PVA in urology including their clinical and research implications.
Methods: Key articles utilizing renal PVA to improve clinical care and facilitate urologic research were reviewed with special emphasis on take-home points of clinical relevance and their contributions to progress in the field.
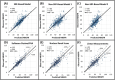
Results: There have been considerable advances in renal PVA over the past 15 years, which is now established as a reference standard for the prediction of functional outcomes after renal surgery. PVA provides improved accuracy when compared to NRS-based estimates or non-SRF-based algorithms. PVA can be performed in minutes using routine preoperative cross-sectional imaging and can be readily applied at the point of care. Additionally, PVA has important research applications, enabling the precise study of the determinants of functional recovery after partial nephrectomy, which can affect surgical approaches to this procedure.
Conclusions: Despite the wide availability of PVA, primarily for use in renal transplantation, it has not been widely implemented for other urologic purposes at most centres. Our hope is that this narrative review will increase PVA utilization in urology and facilitate further progress in the field.
Keywords: functional recovery; kidney cancer; new baseline glomerular filtration rate; parenchymal volume analysis; partial nephrectomy; radical nephrectomy; split renal function.
© 2025 The Author(s). BJUI Compass published by John Wiley & Sons Ltd on behalf of BJU International Company.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Optimizing prediction of new-baseline glomerular filtration rate after radical nephrectomy: are algorithms really necessary?Int Urol Nephrol. 2022 Oct;54(10):2537-2545. doi: 10.1007/s11255-022-03298-y. Epub 2022 Jul 17. Int Urol Nephrol. 2022. PMID: 35842890
-
A Split Renal Function-Based Approach for Predicting New Baseline Glomerular Filtration Rate After Radical Nephroureterectomy.Urol Pract. 2025 May 5:101097UPJ0000000000000826. doi: 10.1097/UPJ.0000000000000826. Online ahead of print. Urol Pract. 2025. PMID: 40323105
-
Limitations of Parenchymal Volume Analysis for Estimating Split Renal Function and New Baseline Glomerular Filtration Rate After Radical Nephrectomy.J Urol. 2024 Jun;211(6):775-783. doi: 10.1097/JU.0000000000003903. Epub 2024 Mar 8. J Urol. 2024. PMID: 38457776
-
Decline in renal function after partial nephrectomy: etiology and prevention.J Urol. 2015 Jun;193(6):1889-98. doi: 10.1016/j.juro.2015.01.093. Epub 2015 Jan 29. J Urol. 2015. PMID: 25637858 Review.
-
Chronic Kidney Disease and Kidney Cancer Surgery: New Perspectives.J Urol. 2020 Mar;203(3):475-485. doi: 10.1097/JU.0000000000000326. Epub 2019 May 7. J Urol. 2020. PMID: 31063051 Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
