Identifying pathological myopia associated genes with GenePlexus in protein-protein interaction network
- PMID: 40110040
- PMCID: PMC11919901
- DOI: 10.3389/fgene.2025.1533567
Identifying pathological myopia associated genes with GenePlexus in protein-protein interaction network
Abstract
Introduction: Pathological myopia, a severe form of myopia, is characterized by an extreme elongation of the eyeball, leading to various vision-threatening complications. It is broadly classified into two primary types: high myopia, which primarily involves an excessive axial length of the eye with potential for reversible vision loss, and degenerative myopia, associated with progressive and irreversible retinal damage.
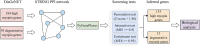
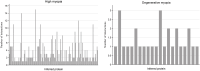
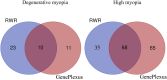
Methods: Leveraging data from DisGeNET, reporting 184 genes linked to high myopia and 39 genes associated with degenerative myopia, we employed the GenePlexus methodology in conjunction with screening tests to further explore the genetic landscape of pathological myopia.
Results and discussion: Our comprehensive analysis resulted in the discovery of 21 new genes associated with degenerative myopia and 133 genes linked to high myopia with significant confidence. Among these findings, genes such as ADCY4, a regulator of the cAMP pathway, were functionally linked to high myopia, while THBS1, involved in collagen degradation, was closely associated with the pathophysiology of degenerative myopia. These previously unreported genes play crucial roles in the underlying mechanisms of pathological myopia, thereby emphasizing the complexity and multifactorial nature of this condition. The importance of our study resides in the uncovering of new genetic associations with pathological myopia, the provision of potential biomarkers for early screening, and the identification of therapeutic targets.
Keywords: DisGeNET; GenePlexus; degenerative myopia; disease gene; high myopia; pathological myopia.
Copyright © 2025 Luo, Wang, Liu, Huang, Lu and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Interventions to slow progression of myopia in children.Cochrane Database Syst Rev. 2020 Jan 13;1(1):CD004916. doi: 10.1002/14651858.CD004916.pub4. Cochrane Database Syst Rev. 2020. PMID: 31930781 Free PMC article.
-
Continued Increase of Axial Length and Its Risk Factors in Adults With High Myopia.JAMA Ophthalmol. 2021 Oct 1;139(10):1096-1103. doi: 10.1001/jamaophthalmol.2021.3303. JAMA Ophthalmol. 2021. PMID: 34436537 Free PMC article.
-
Myopic maculopathy: Current status and proposal for a new classification and grading system (ATN).Prog Retin Eye Res. 2019 Mar;69:80-115. doi: 10.1016/j.preteyeres.2018.10.005. Epub 2018 Nov 1. Prog Retin Eye Res. 2019. PMID: 30391362 Review.
-
Prevalence and Associations of Posterior Segment Manifestations in a Cohort of Egyptian Patients with Pathological Myopia.Curr Eye Res. 2019 Sep;44(9):955-962. doi: 10.1080/02713683.2019.1606252. Epub 2019 Apr 30. Curr Eye Res. 2019. PMID: 30964360
-
[Analysis of the treatment effect of posterior scleral reinforcement on pathological myopia].Zhonghua Yan Ke Za Zhi. 2021 Dec 11;57(12):952-957. doi: 10.3760/cma.j.cn112142-20210707-00324. Zhonghua Yan Ke Za Zhi. 2021. PMID: 34865456 Chinese.
Cited by
-
Early Peripapillary Choroidal Thinning during Myopia Development: A Potential Biomarker for Progressive Myopia Identified in Tree Shrews.Invest Ophthalmol Vis Sci. 2025 Aug 1;66(11):11. doi: 10.1167/iovs.66.11.11. Invest Ophthalmol Vis Sci. 2025. PMID: 40767445 Free PMC article.
References
-
- Akiyama H., Chaboissier M.-C., Martin J. F., Schedl A., De Crombrugghe B. (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes and Dev. 16 (21), 2813–2828. 10.1101/gad.1017802 - DOI - PMC - PubMed
-
- Chen L., Zhang Y. H., Zheng M., Huang T., Cai Y. D. (2016). Identification of compound-protein interactions through the analysis of gene ontology, KEGG enrichment for proteins and molecular fragments of compounds. Mol. Genet. genomics MGG. 291 (6), 2065–2079. 10.1007/s00438-016-1240-x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

