Expression quantitative trait loci associated with performance traits, blood biochemical parameters, and cytokine profile in pigs
- PMID: 40110047
- PMCID: PMC11919875
- DOI: 10.3389/fgene.2025.1533424
Expression quantitative trait loci associated with performance traits, blood biochemical parameters, and cytokine profile in pigs
Abstract
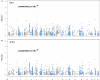
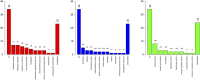
Identifying expression Quantitative Trait Loci (eQTL) and functional candidate variants associated with blood biochemical parameters can contribute to the understanding of genetic mechanisms underlying phenotypic variation in complex traits in pigs. We identified eQTLs through gene expression levels in muscle and liver tissues of Large White pigs. The identified eQTL were then tested for association with biochemical parameters, cytokine profiles, and performance traits of pigs. A total of 41,759 SNPs and 15,093 and 15,516 expression gene levels from muscle and liver tissues, respectively, enabled the identification of 1,199 eQTL. The eQTL identified related the SNP rs345667860 as significantly associated with interleukin-6 and interleukin-18 in liver tissue, while the rs695637860 SNP was associated with aspartate aminotransferase and interleukin-6, and rs337362164 was associated with high-density lipoprotein of the blood serum. In conclusion, the identification of three eQTL significantly associated with aspartate aminotransferase and cytokine levels in both serum and liver tissues suggests a potential role for these variants in modulating immune function and overall health in production pigs. Further research is needed to validate these findings and explore their potential for improving pig health and productivity.
Keywords: GWAS; blood serum indicators; cytokine profile; eQTL; gene expression; inflammatory process; pig; swine.
Copyright © 2025 Freitas, Brito, Silva-Vignato, Nery Ciconello, Almeida and Cesar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Almeida V. V., Silva J. P. M., Schinckel A. P., Meira A. N., Moreira G. C. M., Gomes J. D., et al. (2021). Effects of increasing dietary oil inclusion from different sources on growth performance, carcass and meat quality traits, and fatty acid profile in genetically lean immunocastrated male pigs. Livest. Sci. 248, 104515. 10.1016/j.livsci.2021.104515 - DOI
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources

