Experiences from dual genome next-generation sequencing panel testing for mitochondrial disorders: a comprehensive molecular diagnosis
- PMID: 40110048
- PMCID: PMC11920145
- DOI: 10.3389/fgene.2025.1488956
Experiences from dual genome next-generation sequencing panel testing for mitochondrial disorders: a comprehensive molecular diagnosis
Abstract
Introduction: The molecular diagnosis of mitochondrial disorders is complicated by phenotypic variability, genetic heterogeneity, and the complexity of mitochondrial heteroplasmy. Next-generation sequencing (NGS) of the mitochondrial genome in combination with a targeted panel of nuclear genes associated with mitochondrial disease provides the highest likelihood of obtaining a comprehensive molecular diagnosis. To assess the clinical utility of this approach, we describe the results from a retrospective review of patients having dual genome panel testing for mitochondrial disease.
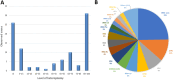
Methods: Dual genome panel testing by NGS was performed on a cohort of 1,509 unrelated affected individuals with suspected mitochondrial disorders. This test included 163 nuclear genes associated with mitochondrial diseases and the entire mitochondrial genome. A retrospective review was performed to evaluate diagnostic yield, disease-gene contributions, and heteroplasmy levels of pathogenic/likely pathogenic (P/LP) mitochondrial DNA (mtDNA) variants.
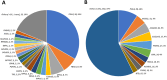
Results: The overall diagnostic yield was 14.6%, with 7.7% from the nuclear genome and 6.9% from the mtDNA genome. P/LP variants in nuclear genes were enriched in both well-established genes (e.g., POLG) and more recently described genes (e.g., FBXL4), highlighting the importance of keeping the panel design updated.
Conclusion: Variants in nuclear and mitochondrial genomes equally contributed to a 14.6% diagnostic yield in this patient cohort. Dual genome NGS testing provides a comprehensive framework for diagnosing mitochondrial disorders, offering clinical utility that can be considered as first-tier approach compared to single genome testing. Characterizing disease-causing genes, variants, and mtDNA heteroplasmy enhances understanding of mitochondrial disorders. Testing alternative tissues can further increase diagnostic yield.
Keywords: NGS; dual-genome; functional group analysis; heteroplasmy; mitochondria.
Copyright © 2025 Gorman, Dai, Feng, Craigen, Chen, Xia, Meng, Liu, Rigobello, Neogi, Eng and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Mitochondrial and nuclear disease panel (Mito-aND-Panel): Combined sequencing of mitochondrial and nuclear DNA by a cost-effective and sensitive NGS-based method.Mol Genet Genomic Med. 2018 Nov;6(6):1188-1198. doi: 10.1002/mgg3.500. Epub 2018 Nov 8. Mol Genet Genomic Med. 2018. PMID: 30406974 Free PMC article.
-
Advanced approach for comprehensive mtDNA genome testing in mitochondrial disease.Mol Genet Metab. 2022 Jan;135(1):93-101. doi: 10.1016/j.ymgme.2021.12.006. Epub 2021 Dec 18. Mol Genet Metab. 2022. PMID: 34969639 Free PMC article.
-
Transition to next generation analysis of the whole mitochondrial genome: a summary of molecular defects.Hum Mutat. 2013 Jun;34(6):882-93. doi: 10.1002/humu.22307. Epub 2013 Apr 2. Hum Mutat. 2013. PMID: 23463613
-
Bioinformatics Tools and Databases to Assess the Pathogenicity of Mitochondrial DNA Variants in the Field of Next Generation Sequencing.Front Genet. 2018 Dec 11;9:632. doi: 10.3389/fgene.2018.00632. eCollection 2018. Front Genet. 2018. PMID: 30619459 Free PMC article. Review.
-
Next generation molecular diagnosis of mitochondrial disorders.Mitochondrion. 2013 Jul;13(4):379-87. doi: 10.1016/j.mito.2013.02.001. Epub 2013 Mar 6. Mitochondrion. 2013. PMID: 23473862 Review.
References
-
- Abicht A., Scharf F., Kleinle S., Schön U., Holinski-Feder E., Horvath R., et al. (2018). Mitochondrial and nuclear disease panel (Mito-aND-Panel): combined sequencing of mitochondrial and nuclear DNA by a cost-effective and sensitive NGS-based method. Mol. Genet. Genomic Med. 6, 1188–1198. 10.1002/mgg3.500 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

