Transcriptional and microbial profile of gastric cancer patients infected with Epstein-Barr virus
- PMID: 40110195
- PMCID: PMC11919665
- DOI: 10.3389/fonc.2025.1530430
Transcriptional and microbial profile of gastric cancer patients infected with Epstein-Barr virus
Abstract
Introduction: Gastric cancer (GC), which has low survival rates and high mortality, is a major concern, particularly in Asia and South America, with over one million annual cases. Epstein-Barr virus (EBV) is recognized as a carcinogen that may trigger gastric carcinogenesis by infecting the stomach epithelium via reactivated B cells, with growing evidence linking it to GC. This study investigates the transcriptional and microbial profiles of EBV-infected versus EBV-non-infected GC patients.
Methods: Using Illumina NextSeq, cDNA libraries were sequenced, and reads were aligned to the human genome and analyzed with DESeq2. Kegg and differential analyses revealed key genes and pathways. Gene sensitivity and specificity were assessed using ROC curves (p < 0.05, AUC > 0.8). Non-aligned reads were used for microbiome analysis with Kraken2 for bacterial identification. Microbial analysis included LDA score, Alpha and Beta diversity metrics, with significance set at p ≤ 0.05. Spearman's correlation between differentially expressed genes (DEGs) and bacteria were also examined.
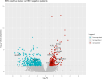
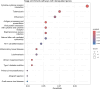
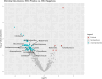
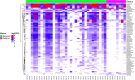
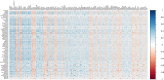
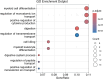
Results: The data revealed a gene expression pattern in EBV-positive gastric cancer, highlighting immune response, inflammation, and cell proliferation genes (e.g., GBP4, ICAM1, IL32, TNFSF10). ROC analysis identified genes with high specificity and sensitivity for discriminating EBV+ gastric cancer, including GBP5, CMKLR1, GM2A and CXCL11 that play pivotal roles in immune response, inflammation, and cancer. Functional enrichment pointed to cytokine-cytokine receptor interactions, antigen processing, and Th17 immune response, emphasizing the role of the tumor microenvironment, shaped by inflammation and immunomodulation, in EBV-associated GC. Microbial analysis revealed changes in the gastric microbiota in EBV+ samples, with a significant reduction in bacterial taxa. The genera Choristoneura and Bartonella were more abundant in EBV+ GC, while more abundant bacteria in EBV- GC included Citrobacter, Acidithiobacillus and Biochmannia. Spearman's correlation showed a strong link between DE bacterial genera and DEGs involved in processes like cell differentiation, cytokine production, digestion, and cell death.
Conclusion: These findings suggest a complex interaction between the host (EBV+ GC) and the microbiota, possibly influencing cancer progression, and offering potential therapeutic targets such as microbiota modulation or gene regulation. Comparing with EBV- samples further highlights the specific impact of EBV and the microbiota on gastric cancer pathogenesis.
Keywords: EBV; carcinogenesis; gastric cancer; gastric microbiome; metatranscriptomics.
Copyright © 2025 Carneiro, Araújo, Da Silva Mourão, Casseb, Demachki, Moreira, Dos Santos, Ishak, Da Costa, Magalhães, Vidal, Burbano and de Assumpção.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Global cancer observatory: cancer today - world . Available online at: https://gco.iarc.who.int/media/globocan/factsheets/populations/900-world... (Accessed November 17, 2024).
-
- Ferlay J, Ervik M, Lam F, Laversanne M, Colombet M, Mery L, et al. . Global Cancer Observatory: Cancer Today - Brazil. International Agency for Research on Cancer. Available online at: https://gco.iarc.who.int/today/ (Accessed November 17, 2024).
LinkOut - more resources
Full Text Sources
Miscellaneous

