Gauge fixing for sequence-function relationships
- PMID: 40111986
- PMCID: PMC11957564
- DOI: 10.1371/journal.pcbi.1012818
Gauge fixing for sequence-function relationships
Abstract
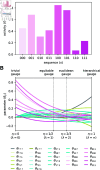
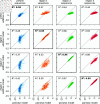
Quantitative models of sequence-function relationships are ubiquitous in computational biology, e.g., for modeling the DNA binding of transcription factors or the fitness landscapes of proteins. Interpreting these models, however, is complicated by the fact that the values of model parameters can often be changed without affecting model predictions. Before the values of model parameters can be meaningfully interpreted, one must remove these degrees of freedom (called "gauge freedoms" in physics) by imposing additional constraints (a process called "fixing the gauge"). However, strategies for fixing the gauge of sequence-function relationships have received little attention. Here we derive an analytically tractable family of gauges for a large class of sequence-function relationships. These gauges are derived in the context of models with all-order interactions, but an important subset of these gauges can be applied to diverse types of models, including additive models, pairwise-interaction models, and models with higher-order interactions. Many commonly used gauges are special cases of gauges within this family. We demonstrate the utility of this family of gauges by showing how different choices of gauge can be used both to explore complex activity landscapes and to reveal simplified models that are approximately correct within localized regions of sequence space. The results provide practical gauge-fixing strategies and demonstrate the utility of gauge-fixing for model exploration and interpretation.
Copyright: © 2025 Posfai et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Update of
-
Gauge fixing for sequence-function relationships.bioRxiv [Preprint]. 2024 Jun 24:2024.05.12.593772. doi: 10.1101/2024.05.12.593772. bioRxiv. 2024. Update in: PLoS Comput Biol. 2025 Mar 20;21(3):e1012818. doi: 10.1371/journal.pcbi.1012818. PMID: 38798671 Free PMC article. Updated. Preprint.
References
-
- Weinberger ED. Fourier and Taylor series on fitness landscapes. Biol Cybern. 1991;65:321–30.
-
- Stadler PF. Landscapes and their correlation functions. J Math Chem. 1996;20:1–45.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

