The nucleosome remodeling and deacetylase-SWItch/sucrose non-fermentable antagonism regulates the coordinated activation of epithelial-to-mesenchymal transition and inflammation in oral cancer
- PMID: 40112045
- PMCID: PMC12229464
- DOI: 10.1093/jnci/djaf065
The nucleosome remodeling and deacetylase-SWItch/sucrose non-fermentable antagonism regulates the coordinated activation of epithelial-to-mesenchymal transition and inflammation in oral cancer
Abstract
Background: Phenotypic plasticity and inflammation, 2 well-established hallmarks of cancer, play key roles in local invasion and distant metastasis by enabling the rapid adaptation of tumor cells to dynamic micro-environmental changes.
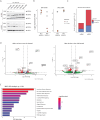
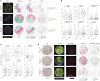
Results: Here, we show that in oral squamous carcinoma cell carcinoma (OSCC), the competition between the Nucleosome Remodeling and Deacetylase (NuRD) and SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complexes plays a pivotal role in regulating both epithelial-mesenchymal plasticity (EMP) and inflammation. By perturbing these complexes, we demonstrated their opposing downstream effects on the inflammatory pathways and EMP regulation. In particular, downregulation of the BRG1-specific SWI/SNF complex deregulates key inflammatory genes, such as TNF-α and IL6, in opposite ways when compared with the loss of CDK2AP1, a key member of the NuRD complex. We showed that CDK2AP1 genetic ablation triggers a pro-inflammatory secretome encompassing several chemokines and cytokines, thus promoting the recruitment of monocytes into the tumor microenvironment (TME). Furthermore, CDK2AP1 deletion stimulates their differentiation into M2-like macrophages, as validated on tumor microarrays from OSCC patient-derived tumor samples. Further analysis of the inverse correlation between CDK2AP1 expression and TME immune infiltration revealed specific downstream effects on the abundance and localization of CD68+ macrophages.
Conclusions: Our study sheds light on the role of chromatin remodeling complexes in OSCC locoregional invasion and highlights the potential of CDK2AP1 and other members of NuRD and SWI/SNF chromatin remodeling complexes as prognostic markers and therapeutic targets.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209-249. https://pubmed.ncbi.nlm.nih.gov/33538338/ - PubMed
-
- Ng JH, Iyer NG, Tan MH, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck. 2017;39:297-304. https://pubmed.ncbi.nlm.nih.gov/27696557/ - PubMed
-
- Hoesseini A, van Leeuwen N, Offerman MPJ, et al. Predicting survival in head and neck cancer: external validation and update of the prognostic model OncologIQ in 2189 patients. Head Neck. 2021;43:2445-2456. https://pubmed.ncbi.nlm.nih.gov/33960553/ - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

