MITD1 is a brain-specific interferon-inducible factor that inhibits flavivirus replication
- PMID: 40112111
- PMCID: PMC11962514
- DOI: 10.1073/pnas.2502064122
MITD1 is a brain-specific interferon-inducible factor that inhibits flavivirus replication
Abstract
West Nile virus (WNV) and Usutu virus (USUV) are closely related mosquito-borne neurotropic flaviviruses that share common transmission cycle and can infect humans. However, while human infections by WNV are widespread, infections by USUV are comparatively less frequent, less severe, and currently limited to Africa and Europe. To identify human host factors that contribute to the pathogenic signatures of these two flaviviruses, we carried out an arrayed expression screen of over 1,300 interferon-stimulated genes (ISGs). Several ISGs known to target flaviviruses, including IFI6, SHFL, and RTP4 were among the strongest hits. Interestingly, we also found MITD1, an ISG with no previously reported antiviral activity, among the strongest hits. We demonstrated that the antiviral activity of MITD1 was not limited to USUV and WNV, since it also inhibited Zika and dengue virus replication. We found MITD1 to interfere with viral RNA replication by sequestering specific endosomal sorting complexes required for transport-III (ESCRT-III) proteins involved in the formation of viral replication factories. MITD1 expression was not increased by type I interferon (IFN-I) in most human cells and mouse tissues that we examined, although WNV and USUV replication was strongly inhibited by IFN-I. Strikingly, MITD1 was induced in the brain of USUV-infected mice and importantly, in human monocyte-derived microglia. Using human microglial-like cells, we confirmed that MITD1 is an essential mediator of the anti-flavivirus activity of IFN-I in these cells. We conclude that MITD1 plays a key role in the cellular defenses against neurotropic flaviviruses.
Keywords: flavivirus; innate immune response; interferon-stimulated genes; microglial cells.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

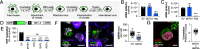

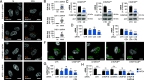

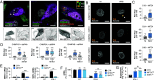
Comment in
-
Tearing down the house of mosquito-transmitted viruses.Proc Natl Acad Sci U S A. 2025 Apr 22;122(16):e2504932122. doi: 10.1073/pnas.2504932122. Epub 2025 Apr 14. Proc Natl Acad Sci U S A. 2025. PMID: 40228137 Free PMC article. No abstract available.
References
-
- Mukhopadhyay S., Kuhn R. J., Rossmann M. G., A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3, 13–22 (2005). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

