Practical microenvironment classification in diffuse large B cell lymphoma using digital pathology
- PMID: 40112808
- PMCID: PMC12047489
- DOI: 10.1016/j.xcrm.2025.102030
Practical microenvironment classification in diffuse large B cell lymphoma using digital pathology
Abstract
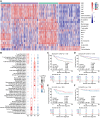
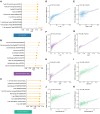
Diffuse large B cell lymphoma (DLBCL) is a heterogeneous B cell neoplasm with variable clinical outcomes influenced by both tumor-derived and lymphoma microenvironment (LME) alterations. A recent transcriptomic study identifies four DLBCL subtypes based on LME characteristics: germinal center (GC)-like, mesenchymal (MS), inflammatory (IN), and depleted (DP). However, integrating this classification into clinical practice remains challenging. Here, we utilize deconvolution methods to assess microenvironment component abundance, establishing an LME classification of DLBCL using immunohistochemistry markers and digital pathology based on CD3, CD8, CD68, PD-L1, and collagen. This staining-based algorithm demonstrates over 80% concordance with transcriptome-based classification. Single-cell sequencing confirms that the immune microenvironments distinguished by this algorithm align with transcriptomic profiles. Significant disparities in overall and progression-free survival are observed among LME subtypes following rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) or R-CHOP with targeted agents (R-CHOP-X) immunochemotherapy. LME subtypes differed from distinct immune escape mechanisms, highlighting specific immunotherapeutic targets and supporting application of this classification in future precision medicine trials.
Keywords: algorithm; cell of origin; diffuse large B cell lymphoma; digital pathology; immune evasion; immunotherapy; lymphoma microenvironment.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Y.-Q.W., S.W., H.-M.Y., P.-P.X., and W.-L.Z. are inventors on a patent application that is based on the work presented herein. W.-L.Z. is a member of the advisory board for Cancer Cell.
Figures





References
-
- Arboe B., Olsen M.H., Gørløv J.S., Duun-Henriksen A.K., Dalton S.O., Johansen C., de Nully Brown P. Treatment intensity and survival in patients with relapsed or refractory diffuse large B-cell lymphoma in Denmark: a real-life population-based study. Clin. Epidemiol. 2019;11:207–216. doi: 10.2147/CLEP.S178003. - DOI - PMC - PubMed
-
- Scott D.W., Wright G.W., Williams P.M., Lih C.J., Walsh W., Jaffe E.S., Rosenwald A., Campo E., Chan W.C., Connors J.M., et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

