Deleterious KOs in the HLA Class I Antigen Processing and Presentation Machinery Induce Distinct Changes in the Immunopeptidome
- PMID: 40113210
- PMCID: PMC12090245
- DOI: 10.1016/j.mcpro.2025.100951
Deleterious KOs in the HLA Class I Antigen Processing and Presentation Machinery Induce Distinct Changes in the Immunopeptidome
Abstract
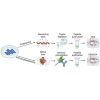
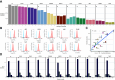
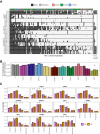
The human leukocyte antigen (HLA) processing and presentation machinery (APPM) is altered in various diseases and in response to drug treatments. Defects in the machinery may change presentation levels or alter the repertoire of presented peptides, globally or in an HLA allele-restricted manner, with direct implications for adaptive immunity. In this study, we investigated the immunopeptidome landscape across a panel of isogenic HAP1 cell line clones, each with a KO of a single gene encoding a key protein in the APPM, including B2M, TAP1, TAP2, TAPBP, IRF2, PDIA3, ERAP1, GANAB, SPPL3, CANX, and CALR. We applied immunopeptidomics and proteomics to assess the successful gene KOs on the protein level, understand how these proteins participate in antigen presentation, and contextualize protein expression and antigen presentation. We validated the absence of the KO proteins in the respective samples and found that knocking-out an APPM component leads to the loss of peptide subsets that are normally presented on the control wildtype cells. We assessed the immunopeptidomes qualitatively and quantitatively, considering factors like peptide diversity, peptide length distribution, and binding affinity to the endogenously expressed HLA alleles in HAP1 cells. We demonstrated prominent HLA allele-restricted alterations in several KO conditions. The absence of CALR, CANX, and TAP1 led to significant changes in HLA allele-specific presentation levels. Overall, this work represents the first systematic analysis of how the absence of individual APPM components, knocked out in a single cell line under controlled conditions, affects the immunopeptidome. This approach could facilitate the creation of predictive tools capable of prioritizing HLA-bound peptides likely to be presented when presentation defects occur, such as in cancer and viral infections.
Keywords: antigen processing and presentation machinery; human leukocyte antigen class I; immunopeptidomics.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest R. S. currently works at Neogene Therapeutics, a member of the AstraZeneca Group. All other authors declare no competing interests.
Figures






References
-
- Pishesha N., Harmand T.J., Ploegh H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022;22:751–764. - PubMed
-
- Jongsma M.L.M., Neefjes J., Spaapen R.M. Playing hide and seek: tumor cells in control of MHC class I antigen presentation. Mol. Immunol. 2021;136:36–44. - PubMed
-
- Jhunjhunwala S., Hammer C., Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer. 2021;21:298–312. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

