Assessing the potential for non-digestible carbohydrates toward mitigating adverse effects of antibiotics on microbiota composition and activity in an in vitro colon model of the weaning infant
- PMID: 40113239
- PMCID: PMC11963755
- DOI: 10.1093/femsec/fiaf028
Assessing the potential for non-digestible carbohydrates toward mitigating adverse effects of antibiotics on microbiota composition and activity in an in vitro colon model of the weaning infant
Abstract
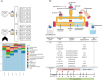
Environmental factors like diet and antibiotics modulate the gut microbiota in early life. During weaning, gut microbiota progressively diversifies through exposure to non-digestible carbohydrates (NDCs) from diet, while antibiotic perturbations might disrupt this process. Supplementing an infant's diet with prebiotic NDCs may mitigate the adverse effects of antibiotics on gut microbiota development. This study evaluated the influence of supplementation with 2-fucosyllactose (2'-FL), galacto-oligosaccharides (GOS), or isomalto/malto-polysaccharides containing 87% of α(1→6) linkages (IMMP-87), on the recovery of antibiotic-perturbed microbiota. The TIM-2 in vitro colon model inoculated with fecal microbiota of 9-month-old infants was used to simulate the colon of weaning infants exposed to the antibiotics amoxicillin/clavulanate or azithromycin. Both antibiotics induced changes in microbiota composition, with no signs of recovery in azithromycin-treated microbiota within 72 h. Moreover, antibiotic exposure affected microbiota activity, indicated by a low valerate production, and azithromycin treatment was associated with increased succinate production. The IMMP-87 supplementation promoted the compositional recovery of amoxicillin/clavulanate-perturbed microbiota, associated with the recovery of Ruminococcus, Ruminococcus gauvreauii group, and Holdemanella. NDC supplementation did not influence compositional recovery of azithromycin-treated microbiota. Irrespective of antibiotic exposure, supplementation with 2'-FL, GOS, or IMMP-87 enhanced microbiota activity by increasing short-chain fatty acids production (acetate, propionate, and butyrate).
Keywords: 2′-FL; GOS; IMMP; amoxicillin/clavulanate; azithromycin; gut bacteria.
© The Author(s) 2025. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
A.N. is employed by FrieslandCampina and H.L. is employed by Avebe. The remaining authors declare no competing interests.
Figures







References
-
- Apprill A, McNally S, Parsons R et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–37.
-
- Barnett DJ, Arts IC, Penders J. microViz: an R package for microbiome data visualization and statistics. J Open Source Softw. 2021;6:3201.
-
- Bower CE, Holm-Hansen T. A salicylate–hypochlorite method for determining ammonia in seawater. Can J Fish Aquat. 1980;37:794–8.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

