Optical genome mapping enables accurate testing of large repeat expansions
- PMID: 40113266
- PMCID: PMC12047237
- DOI: 10.1101/gr.279491.124
Optical genome mapping enables accurate testing of large repeat expansions
Abstract
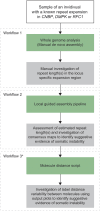
Short tandem repeats (STRs) are common variations in human genomes that frequently expand or contract, causing genetic disorders, mainly when expanded. Traditional diagnostic methods for identifying these expansions, such as repeat-primed PCR and Southern blotting, are often labor-intensive, locus-specific, and are unable to precisely determine long repeat expansions. Sequencing-based methods, although capable of genome-wide detection, are limited by inaccuracy (short-read technologies) and high associated costs (long-read technologies). This study evaluated optical genome mapping (OGM) as an efficient, accurate approach for measuring STR lengths and assessing somatic stability in 85 samples with known pathogenic repeat expansions in DMPK, CNBP, and RFC1, causing myotonic dystrophy types 1 and 2 and cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS), respectively. Three workflows-manual de novo assembly, local guided assembly (local-GA), and a molecule distance script-were applied, of which the latter two were developed as part of this study to assess the repeat sizes and somatic repeat stability. OGM successfully identified 84/85 (98.8%) of the pathogenic expansions, distinguishing between wild-type and expanded alleles or between two expanded alleles in recessive cases, with greater accuracy than standard of care (SOC) for long repeats and no apparent upper size limit. Notably, OGM detected somatic instability in a subset of DMPK, CNBP, and RFC1 samples. These findings suggest OGM could advance diagnostic accuracy for large repeat expansions, providing a more comprehensive genome-wide assay for repeat expansion disorders by measuring exact repeat lengths and somatic instability across multiple loci simultaneously.
© 2025 van der Sanden et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Alfano M, De Antoni L, Centofanti F, Visconti VV, Maestri S, Degli Esposti C, Massa R, D'Apice MR, Novelli G, Delledonne M, et al. 2022. Characterization of full-length CNBP expanded alleles in myotonic dystrophy type 2 patients by Cas9-mediated enrichment and nanopore sequencing. Elife 11: e80229. 10.7554/eLife.80229 - DOI - PMC - PubMed
-
- Barseghyan H, Pang AWC, Zhang Y, Sahajpal NS, Delpu Y, Lai C-YJ, Lee J, Tessereau C, Oldakowski M, Kolhe RB, et al. 2022. Neurogenetic variant analysis by optical genome mapping for structural variation detection-balanced genomic rearrangements, copy number variants, and repeat expansions/contractions. In Genomic structural variants in nervous system disorders (ed. Proukakis C), pp. 155–172. Springer, New York.
-
- Cumming SA, Hamilton MJ, Robb Y, Gregory H, McWilliam C, Cooper A, Adam B, McGhie J, Hamilton G, Herzyk P, et al. 2018. De novo repeat interruptions are associated with reduced somatic instability and mild or absent clinical features in myotonic dystrophy type 1. Eur J Hum Genet 26: 1635–1647. 10.1038/s41431-018-0156-9 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
