PER2 interaction with HSP70 promotes cuproptosis in oral squamous carcinoma cells by decreasing AKT stability
- PMID: 40113747
- PMCID: PMC11926231
- DOI: 10.1038/s41419-025-07523-1
PER2 interaction with HSP70 promotes cuproptosis in oral squamous carcinoma cells by decreasing AKT stability
Abstract
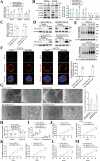
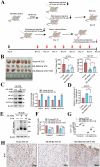
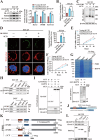
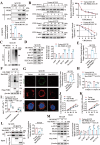
Oral squamous cell carcinoma (OSCC) has a poor prognosis, with unclear mechanisms posing a challenge for the development of effective treatment strategies. Cuproptosis is a novel cell death mode that disrupts mitochondrial metabolism. Clarifying the mechanisms that regulate cuproptosis may provide important new insights to guide OSCC treatment. Here, we found that the biological clock gene Period2 (PER2) was under-expressed in OSCC, with consequent inhibition of cellular cuproptosis, whereas it was overexpression of PER2 in vitro and in vivo and promoted OSCC cellular cuproptosis. Mechanistically, PER2 bound to heat shock protein 70 (HSP70) through its C-terminal domain, subsequently reducing the interaction between HSP70 and AKT and leading to enhanced degradation of AKT ubiquitination, and promoting cuproptosis in OSCC cells by inhibiting the AKT pathway and upregulating DLAT, PDHB, and SLC31A1 expression. Activating transcription factor 3 (ATF3) is an upstream regulator of PER2, that binds to the -807 to -796 bp site of the PER2 promoter. Overexpression of ATF3 in vitro and in vivo is dependent on transcriptional activation of PER2 and promotes cuproptosis in OSCC cells. The anti-tumor effect of ATF3 inducer 1-targeted upregulation of PER2 combined with copper ionophore elesclomol (ES) was found to be significantly enhanced compared with that of monotherapy in an OSCC xenograft model. These findings reveal a critical role of ATF3-dependent regulation of cuproptosis by PER2 in OSCC development, suggesting targeted upregulation of PER2 or ATF3 in combination to induce cuproptosis as a novel strategy to potentially improve the prognosis of OSCC patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The author declares no competing interests. Ethical approval: This study was approved by the Ethics Committee of First Affiliated Hospital of Chongqing Medical University, in accordance with the Declaration of Helsinki. All animal experiments were approved by the Institutional Animal Care and Use committee of Chongqing Medical University.
Figures



References
-
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Bhat GR, Hyole RG, Li J. Head and neck cancer: current challenges and future perspectives. Adv Cancer Res. 2021;152:67–102. - PubMed
-
- Cramer JD, Burtness B, Le QT, Ferris RL. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol. 2019;16:669–83. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

