Targeting PI3K inhibitor resistance in breast cancer with metabolic drugs
- PMID: 40113784
- PMCID: PMC11926384
- DOI: 10.1038/s41392-025-02180-4
Targeting PI3K inhibitor resistance in breast cancer with metabolic drugs
Abstract
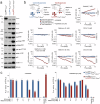
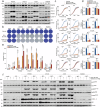
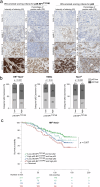
Activating PIK3CA mutations, present in up to 40% of hormone receptor-positive (HR+), human epidermal growth factor receptor 2-negative (Her2-) breast cancer (BC) patients, can be effectively targeted with the alpha isoform-specific PI3K inhibitor Alpelisib. This treatment significantly improves outcomes for HR+, Her2-, and PIK3CA-mutated metastatic BC patients. However, acquired resistance, often due to aberrant activation of the mTOR complex 1 (mTORC1) pathway, remains a significant clinical challenge. Our study, using in vitro and orthotopic xenograft mouse models, demonstrates that constitutively active mTORC1 signaling renders PI3K inhibitor-resistant BC exquisitely sensitive to various drugs targeting cancer metabolism. Mechanistically, mTORC1 suppresses the induction of autophagy during metabolic perturbation, leading to energy stress, a critical depletion of aspartate, and ultimately cell death. Supporting this mechanism, BC cells with CRISPR/Cas9-engineered knockouts of canonical autophagy genes showed similar vulnerability to metabolically active drugs. In BC patients, high mTORC1 activity, indicated by 4E-BP1T37/46 phosphorylation, correlated with p62 accumulation, a sign of impaired autophagy. Together, these markers predicted poor overall survival in multiple BC subgroups. Our findings reveal that aberrant mTORC1 signaling, a common cause of PI3K inhibitor resistance in BC, creates a druggable metabolic vulnerability by suppressing autophagy. Additionally, the combination of 4E-BP1T37/46 phosphorylation and p62 accumulation serves as a biomarker for poor overall survival, suggesting their potential utility in identifying BC patients who may benefit from metabolic therapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.D. received personal fees from Novartis, Roche, MSD Oncology, Daiichi-Sankyo, AstraZeneca, Molecular Health, and Merck, all outside the submitted work. C.D. is cofounder of Sividon Diagnostics. In addition, C.D. has a patent on VMScope digital pathology software with royalties paid; a patent WO2020109570A1—cancer immunotherapy pending; and patents WO2015114146A1 and WO2010076322A1—therapy response issued. P.J. reports research grants and travel expenses from GILEAD Sciences GmbH outside the submitted work. N.H. declares to be a GBG Forschungs GmbH employee. GBG Forschungs GmbH reports financial funding from AstraZeneca and Myriad during the conduct of the study; received funding for research grants from Abbvie, Amgen, AstraZeneca, BMS, Daiichi-Sankyo, Gilead, Molecular Health, Stemline Menarini, Celgene/BMS, Novartis, Pfizer and Roche (paid to the institution); GBG Forschungs GmbH has licensing fees from VMscope GmbH. In addition, GBG Forschungs GmbH has a patent EP21152186.9 pending, a patent EP19808852.8 pending, and a patent EP14153692.0 pending. S.L. declares to be the CEO of the GBG Forschungs GmbH. GBG Forschungs GmbH receives grants and other from Abbvie, grants from AstraZeneca, grant from Amgen, grants from DSI, grants from Gilead, Grants from Molecular Health, grants from Celgene/BMS, grants from Novartis, grants from Pfizer, grants from Roche, grant from Stemline/Menarini, financial support from Astra Zeneca, Gilead, Myriad, Novartis, Pfizer, BMS, Sanofi, Lilly, MSD; Consulting fees from following: AstraZeneca, Abbvie, Amgen, Cellcuity, DSI, Gilead, Novartis, Incyte, Pfizer, BMC, Sanofi, Lilly, MSD, Esai, Exact Science, Relay, Stemline, GSK, BioNtech, Olema, Roche; Payment Honoria: AZ, Amgen, Agendia, DSI, Gilead, Novartis, Pfizer, BMS, Stemline/Menarini, Lilly, MSD, Seagen, Medscape, Pierre Fabre, Roche; Travel support from: DSI, ESMO, SCGCC. S.L. reports leadership in: ESMO, SBGCC, SABCS, and AGO Kommission Mamma, S.L. reports receipt of drug equipment in trials from: Gilead, AZ, Celgene/BMS, Novartis, Pfizer, and Roche. GBG Forschungs GmbH has licensing fees from VMscope GmbH. In addition, GBG Forschungs GmbH has a patent EP21152186.9 pending, a patent EP19808852.8 pending, and a patent EP14153692.0 pending. The other authors declare no conflicts of interest. Ethics approval and consent to participate: Immunohistochemical staining and evaluation of breast cancer patient samples was approved by the Ethics Committee of the University of Marburg (Ethics Opinion No 38/20). All xenograft experiments were performed according to the German animal welfare law and the European legislation for the protection of animals used for scientific purposes (2010/63/EU) and were approved by the regional board (RP Giessen).
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

