High proportions of multidrug-resistant Klebsiella pneumoniae isolates in community-acquired infections, Brazil
- PMID: 40113819
- PMCID: PMC11926192
- DOI: 10.1038/s41598-025-92549-w
High proportions of multidrug-resistant Klebsiella pneumoniae isolates in community-acquired infections, Brazil
Abstract
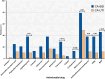
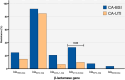
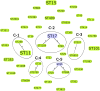
Klebsiella pneumoniae is one of the leading causes of bloodstream (BSI) and urinary tract infections (UTI), but limited data is available regarding community-acquired (CA) infections. This study characterized the clinical aspects of CA-BSI and CA-UTI caused by K. pneumoniae and the molecular features of isolates, including their resistance profiles. Sixty-five isolates (CA-BSI, n = 24; CA-UTI, n = 41) underwent antimicrobial susceptibility testing, β-lactamase and virulence gene assessment, capsular genotyping, and molecular typing. Older age, male gender, and comorbidities, particularly kidney disease, were significantly associated with CA-BSI. The MDR and carbapenem resistance rates for K. pneumoniae from CA infections were 24.6% and 4.6%, respectively. CA-BSI isolates were more antibiotic-resistant and had a higher proportion of ESBL-producing (37.5% versus 9.8%) and MDR isolates (45.8% versus 12.2%) than CA-UTI. The blaCTX-M-like or blaKPC-like genes was found in all ESBL-producing isolates, while blaKPC-like and blaNDM-like were detected exclusively in CA-BSI strains. The isolates' virulence profiles were similar between the groups, although one CA-BSI and two CA-UTI isolates presented hypervirulence biomarkers. A high clonal diversity was observed, with a majority of MDR (81.3%) (ST11, ST15, ST101, ST258, ST307, and ST6852) and hypervirulent (2/3) (ST23 and ST65) isolates being high-risk pandemic clones in humans. Our data highlight the high prevalence of MDR K. pneumoniae in CA infections in Brazil, with CA-BSI showing significant differences in resistance profiles compared to CA-UTI.
Keywords: Klebsiella pneumoniae; Antimicrobial resistance; Bloodstream infections; Community-acquired; High-risk clones; Urinary tract infections.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Devi, L. et al. Increasing prevalence of Escherichia coli and Klebsiella pneumoniae producing CTX-M-type extended-spectrum beta-lactamase, carbapenemase, and NDM-1 in patients from a rural community with community acquired infections: A 3-year study. Int. J. Appl. Basic. Med. Res.10, 156–163. 10.4103/ijabmr.IJABMR_360_19 (2020). - PMC - PubMed
-
- Azevedo, P. A. A. et al. Molecular characterisation of multidrug-resistant Klebsiella pneumoniae belonging to CC258 isolated from outpatients with urinary tract infection in Brazil. J. Glob Antimicrob. Resist.18, 74–79. 10.1016/j.jgar.2019.01.025 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

