Bcor loss promotes Richter transformation of chronic lymphocytic leukemia associated with Notch1 activation in mice
- PMID: 40113912
- PMCID: PMC12055590
- DOI: 10.1038/s41375-025-02557-y
Bcor loss promotes Richter transformation of chronic lymphocytic leukemia associated with Notch1 activation in mice
Abstract
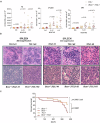
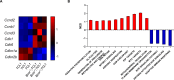
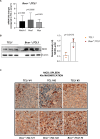
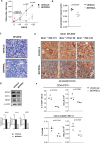
Richter's transformation (RT) is an aggressive lymphoma occurring upon progression from chronic lymphocytic leukemia (CLL). Despite advances in deciphering the RT genetic architecture, the mechanisms driving this disease remain unknown. BCOR disruptive mutations were found in CLL and frequently associated with NOTCH1 aberrations, a common feature in CLL and RT. We engineered mice to knock-out Bcor in B and CLL cells of Eμ-TCL1 mice. Bcor loss resulted in alterations of the B cell compartment and favored CLL transformation into an aggressive lymphoma with reduced survival in Eμ-TCL1 mice. RNA-sequencing demonstrated a molecular signature reminiscent of human RT and implied the involvement of the T cell tumour microenvironment in the disease onset. Bcor deficiency was associated with Notch1 activation in splenic CD19 + CD5+ cells to accelerate Eμ-TCL1 mice lymphoproliferation. Notch1 inhibition progressively reduced circulating CD19+ CD5+ and RT cells infiltrating the spleen of diseased mice with concomitant reduction of PD-1 expressing T cells and improved survival. Our data demonstrated an interplay between the tumour suppressor activity of Bcor and Notch1 in RT pathogenesis with potential for tumour targeting. This model represented a new platform to uncover promising alternatives for this incurable tumour.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All methods were performed in accordance with the relevant guidelines and regulations. The Institutional Animal Care and Use Committee approved all the procedures. Experiments were performed with the approval of the Italian Ministry of Health (authorization n°971/2020-PR and n°253/2024-PR).
Figures


References
-
- Sportoletti P, Baldoni S, Del Papa B, Aureli P, Dorillo E, Ruggeri L, et al. A revised NOTCH1 mutation frequency still impacts survival while the allele burden predicts early progression in chronic lymphocytic leukemia. Leukemia. 2014;28:436–9. - PubMed
MeSH terms
Substances
Grants and funding
- IG 2024 - ID. 30768/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- IG 2018 - ID 21352/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- ID.23928/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- ID.26617/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

