Lack of AtMC1 catalytic activity triggers autoimmunity dependent on NLR stability
- PMID: 40113992
- PMCID: PMC12069761
- DOI: 10.1038/s44319-025-00426-4
Lack of AtMC1 catalytic activity triggers autoimmunity dependent on NLR stability
Abstract
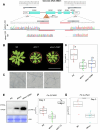
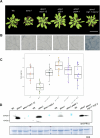
Plants utilize cell surface-localized pattern recognition receptors (PRRs) and intracellular nucleotide-binding leucine-rich repeat (NLR) receptors to detect non-self and elicit robust immune responses. Fine-tuning the homeostasis of these receptors is critical to prevent their hyperactivation. Here, we show that Arabidopsis plants lacking metacaspase 1 (AtMC1) display autoimmunity dependent on immune signalling components downstream of NLR and PRR activation. Overexpression of a catalytically inactive AtMC1 in an atmc1 background triggers severe autoimmunity partially dependent on the same immune signalling components. Overexpression of the E3 ligase SNIPER1, a master regulator of NLR homeostasis, fully reverts the AtMC1-dependent autoimmunity phenotype, inferring that a broad defect in NLR turnover may underlie the severe phenotype observed. Catalytically inactive AtMC1 localizes to punctate structures that are degraded through autophagy. Considering also previous evidence on the proteostatic functions of AtMC1, we speculate that Wt AtMC1 may either directly or indirectly control NLR protein levels, thereby preventing autoimmunity.
Keywords: Autoimmunity; Autophagy; Condensates; Metacaspases; Proteostasis.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures













References
-
- Asqui Lema S, Vercammen D, Serrano I, Valls M, Rivas S, Van Breusegem F, Conlon FL, Dangl JL, Coll NS (2018) AtSERPIN1 is an inhibitor of the metacaspase AtMC1‐mediated cell death and autocatalytic processing in planta. New Phytol 218:1156–1166 - PubMed
-
- Bi G, Su M, Li N, Liang Y, Dang S, Xu J, Hu M, Wang J, Zou M, Deng Y et al (2021) The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184:3528–3541.e12 - PubMed
-
- Bomblies K, Weigel D (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8:382–393 - PubMed
MeSH terms
Substances
Grants and funding
- PID2022-136922NB-I00,PID2019-108595RB-I00,TED2021-131457B-I00/Ministerio de Ciencia e Innovación (MCIN)
- BES-2017-080210/Ministerio de Ciencia e Innovación (MCIN)
- PREP2022-000557/Ministerio de Ciencia e Innovación (MCIN)
- CEX2019-000902-S/Ministerio de Ciencia e Innovación (MCIN)
- FJC2021-046667/Ministerio de Ciencia e Innovación (MCIN)
- MCIN/AEI/10.13039/501100011033/Ministerio de Ciencia e Innovación (MCIN)
- FPU19/03778/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- 945043/EC | H2020 | PRIORITY 'Excellent science' | H2020 Marie Skłodowska-Curie Actions (MSCA)
- rePLANT-GA101081581/EC | H2020 | PRIORITY 'Excellent science' | H2020 Marie Skłodowska-Curie Actions (MSCA)
- GA#101068121/EC | H2020 | PRIORITY 'Excellent science' | H2020 Marie Skłodowska-Curie Actions (MSCA)
- GA#945043/EC | H2020 | PRIORITY 'Excellent science' | H2020 Marie Skłodowska-Curie Actions (MSCA)
- CRC1101 D09/Deutsche Forschungsgemeinschaft (DFG)
- helperless plant/Reinhard-Frank Stiftung
- No grant number/Centres de Recerca de Catalunya (CERCA)
LinkOut - more resources
Full Text Sources

