Acquired resistance to immune checkpoint therapy is caused by glycoprotein non-metastatic melanoma protein B signal cascade
- PMID: 40114009
- PMCID: PMC11926377
- DOI: 10.1038/s43856-025-00786-x
Acquired resistance to immune checkpoint therapy is caused by glycoprotein non-metastatic melanoma protein B signal cascade
Abstract
Background: Acquired resistance (AR) is a major limitation of immune checkpoint inhibitor (ICI) therapy when treating renal cell carcinoma (RCC). Understanding who will get AR is currently unknown. We hypothesized the T-cell-inhibitory glycoprotein non-metastatic melanoma protein B (GPNMB) to be a prognostic marker for patients with AR.
Methods: Alongside other markers, GPNMB was measured in the blood of RCC patients (n = 39) several times after starting ICI treatment and analyzed for association with Response Evaluation Criteria in Solid Tumors (RECIST) tumor response. To better understand the role of GPNMB in AR, we created an ICI-resistant RenCa mouse kidney cancer model by repeatedly selecting the largest tumors that grew in ICI-treated mice.
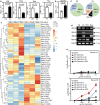
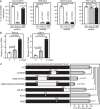
Results: Here we show that among patients who positively respond to ICI, a subset of patients (n = 9) acquire resistance within 2 years that coincides with an increase in serum GPNMB. Our mouse model recapitulates this elevation in GPNMB at the onset of AR which is triggered by cytoplasmic motif signaling via the Programmed cell death ligand 1 (PDL1) receptor that is known to protect tumors from Interferon-gamma (IFN-γ) cytotoxicity. This PDL1-induced signal leads to upregulation of the SRY-box transcription factor 10 (SOX10), dysregulation of the microphthalmia-associated transcription factor (MITF) pathway, and overexpression of GPNMB. Indeed, activation of SOX10-MITF signaling is present in plasma cell-free RNA from RCC patients who develop AR.
Conclusions: Elevation of the SOX10-MITF-GPNMB signal cascade via the PDL1 receptor leads to AR. Therefore, GPNMB can be both a prognosticator of and a potential treatment target for overcoming AR to ICI treatment in RCC.
Plain language summary
Immune checkpoint inhibitors (ICI) are a type of cancer treatment that helps the immune system kill cancer cells. However, over half of people with kidney cancer who initially see a benefit following ICI treatment find the drug stops working for them, with their cancer acquiring resistance to this treatment. In this study, we investigated if a protein known as glycoprotein non-metastatic melanoma protein B (GPNMB) increases in cancers that do not respond to ICI therapy. We find that elevated GPNMB levels coincide with the onset of treatment resistance to ICI therapy in both humans and a mouse model of kidney cancer. Therefore, measuring GPNMB levels during ICI therapy can inform if a patient will become resistant to treatment. Additionally, GPNMB may serve as a potential target for overcoming such resistance.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Hammers, H. Immunotherapy in kidney cancer: the past, present, and future. Curr. Opin. Urol.26, 543–547 (2016). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

