CircPVT1 weakens miR-33a-5p unleashing the c-MYC/GLS1 metabolic axis in breast cancer
- PMID: 40114244
- PMCID: PMC11924866
- DOI: 10.1186/s13046-025-03355-1
CircPVT1 weakens miR-33a-5p unleashing the c-MYC/GLS1 metabolic axis in breast cancer
Abstract
Background: Altered metabolism is one of the cancer hallmarks. The role of circRNAs in cancer metabolism is poorly studied. Specifically, the impact of circPVT1, a well-known oncogenic circRNA on triple negative breast cancer metabolism is mechanistically underexplored.
Methods: The clinical significance of circPVT1 expression levels was assessed in human breast cancer samples using digital PCR and the cancer genome atlas (TCGA) dataset. The oncogenic activity of circPVT1 was assessed in TNBC cell lines and in MCF-10 A breast cell line by either ectopic expression or depletion of circPVT1 molecule. CircPVT1 mediated metabolic perturbation was assessed by 1 H-NMR spectroscopy metabolic profiling. The binding of circPVT1 to miR-33a-5p and c-Myc recruitment onto the Glutaminase gene promoter were assessed by RNA immunoprecipitation and chromatin immunoprecipitation assays, respectively. The circPVT1/miR-33a-5p/Myc/GLS1 axis was functionally validated in breast cancer patients derived organoids. The viability of 2D and PDO cell models was assessed by ATP light assay and Opera Phenix plus high content screening.
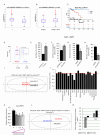
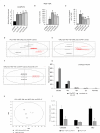
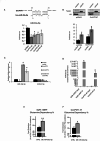
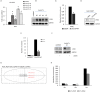
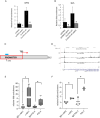
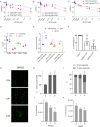
Results: We initially found that the expression of circPVT1 was significantly higher in tumoral tissues than in non-tumoral breast tissues. Basal like breast cancer patients with higher levels of circPVT1 exhibited shorter disease-free survival compared to those with lower expression. CircPVT1 ectopic expression rendered fully transformed MCF-10 A immortalized breast cells and increased tumorigenicity of TNBC cell lines. Depletion of endogenous circPVT1 reduced tumorigenicity of SUM-159PT and MDA-MB-468 cells. 1 H-NMR spectroscopy metabolic profiling of circPVT1 depleted breast cancer cell lines revealed reduced glycolysis and glutaminolitic fluxes. Conversely, MCF-10 A cells stably overexpressing circPVT1 exhibited increased glutaminolysis. Mechanistically, circPVT1 sponges miR-33a-5p, a well know metabolic microRNA, which in turn releases c-MYC activity promoting transcriptionally glutaminase. This activity facilitates the conversion of glutamine to glutamate. CircPVT1 depletion synergizes with GLS1 inhibitors BPTES or CB839 to reduce cell viability of breast cancer cell lines and breast cancer-derived organoids.
Conclusions: In aggregate, our findings unveil the circPVT1/miR-33a-5p/Myc/GLS1 axis as a pro-tumorigenic metabolic event sustaining breast cancer transformation with potential therapeutic implications.
Keywords: Breast cancer; MYC; Metabolism; Non-coding RNAs; Patients derived organoids.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Generation of patient-derived organoids from breast cancer was approved by institutional review board of Regina Elena National Cancer Institute and appropriate regulatory authorities (approval no. IFO 1270/19). All patients signed an informed consent. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
-
- Schmid P, Cortes J, Pusztai L, McArthur H, Kummel S, Bergh J, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020;382:810–21. - PubMed
-
- Chen X, Zhou M, Yant L, Huang C. Circular RNA in disease: basic properties and biomedical relevance. Wiley Interdiscip Rev RNA. 2022;13:e1723. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

