TBK1 is a signaling hub in coordinating stress-adaptive mechanisms in head and neck cancer progression
- PMID: 40114316
- PMCID: PMC12282999
- DOI: 10.1080/15548627.2025.2481661
TBK1 is a signaling hub in coordinating stress-adaptive mechanisms in head and neck cancer progression
Abstract
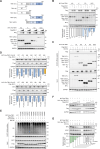
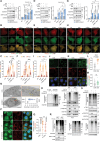
Tumorigenesis is closely linked to the ability of cancer cells to activate stress-adaptive mechanisms in response to various cellular stressors. Stress granules (SGs) play a crucial role in promoting cancer cell survival, invasion, and treatment resistance, and influence tumor immune escape by protecting essential mRNAs involved in cell metabolism, signaling, and stress responses. TBK1 (TANK binding kinase 1) functions in antiviral innate immunity, cell survival, and proliferation in both the tumor microenvironment and tumor cells. Here, we report that MUL1 loss results in the hyperactivation of TBK1 in both HNC cells and tissues. Mechanistically, under proteotoxic stress induced by proteasomal inhibition, HSP90 inhibition, or Ub+ stress, MUL1 promotes the degradation of active TBK1 through K48-linked ubiquitination at lysine 584. Furthermore, TBK1 facilitates autophagosome-lysosome fusion and phosphorylates SQSTM1, regulating selective macroautophagic/autophagic clearance in HNC cells. TBK1 is required for SG formation and cellular protection. Moreover, we found that MAP1LC3B is partially localized within SGs. TBK1 depletion enhances the sensitivity of HNC cells to cisplatin-induced cell death. GSK8612, a novel TBK1 inhibitor, significantly inhibits HNC tumorigenesis in xenografts. In summary, our study reveals that TBK1 facilitates the rapid removal of ubiquitinated proteins within the cell through protective autophagy under stress conditions and assists SG formation through the use of the autophagy machinery. These findings highlight the potential of TBK1 as a therapeutic target in HNC treatment.Abbreviations: ALP: autophagy-lysosomal pathway; AMBRA1: autophagy and beclin 1 regulator 1; BaF: bafilomycin A1; CC: coiled-coil; CD274/PDL-1: CD274 molecule; CHX: cycloheximide; CQ: chloroquine; DNP: dinitrophenol; EGFR: epidermal growth factor receptor; ESCC: esophageal squamous cell carcinoma; G3BP1: G3BP stress granule assembly factor 1; HNC: head and neck cancer; HPV: human papillomavirus; IFN: interferon; IGFBP3: insulin like growth factor binding protein 3; IRF: interferon-regulatory factor 3; KO: knockout; LAMP1: lysosomal associated membrane protein 1; MAP1LC3B: microtubule associated protein 1 light chain 3 beta; NPC: nasopharyngeal carcinoma; PABP: poly(A) binding protein; PI: proteasome inhibitor; PQC: protein quality control; PROTAC: proteolysis-targeting chimera; PURA/PURα: purine rich element binding protein A; RIGI: RNA sensor RIG-I; SD: standard deviation; SG: stress granule; SQSTM1: sequestosome 1; STING1: stimulator of interferon response cGAMP interactor 1; TBK1: TANK binding kinase 1; UPS: ubiquitin-proteasome system; USP10: ubiquitin specific peptidase 10; VCP: valosin containing protein; VHL: von Hippel-Lindau tumor suppressor; WT: wild type.
Keywords: Autophagic flux; GSK8612; MUL1; TBK1; head and neck cancer; stress granule formation.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
