Unveiling the Genetic Landscape of Staphylococcus aureus Isolated From Hospital Wastewaters: Emergence of Hypervirulent CC8 Strains in Tehran, Iran
- PMID: 40114671
- PMCID: PMC11925629
- DOI: 10.1155/ijm/5458315
Unveiling the Genetic Landscape of Staphylococcus aureus Isolated From Hospital Wastewaters: Emergence of Hypervirulent CC8 Strains in Tehran, Iran
Abstract
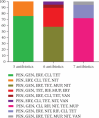
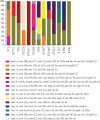
Objective(s): Multidrug-resistant bacteria and priority pathogens, including MRSA, are frequently found in hospital wastewaters. It is crucial to investigate the genetic diversity, biofilm formation, and virulence analysis of Staphylococcus aureus isolated from hospital wastewaters. Materials and Methods: In this cross-sectional study, 70 S. aureus isolated from hospital wastewaters were subjected to characterization through antimicrobial susceptibility tests, biofilm formation, multilocus sequence typing (MLST), and PCR analysis for detecting resistance (mecA, mecC, vanA, vanB, mupB, mupA, msr(A), msr(B), erm(A), erm(B), erm(C), tet(M), ant (4')-Ia, aac (6')-Ie/aph (2″), and aph (3')-IIIa) and virulence genes (eta, etb, pvl, and tst). Results: Our results showed that 55.7%, 31.4%, and 12.9% of isolates were classified as strong, intermediate, and weak biofilm-forming strains, respectively. Our result revealed that about three-quarters of isolates harbored mecA (100%), ant (4')-Ia (100%), tet(M) (92.9%), erm(B) (80%), and msr(A) (74.3%) resistance genes. MLST revealed that the 70 isolates belonged to five clonal complexes, including CC8 (52.9%), followed by CC30 (15.7%), CC5 (14.3%), CC1 (11.4%), and CC22 (5.7%). The vast majority of S. aureus isolates belonged to CC8/ST239-MRSA (21.5%). Among the 39 strong biofilm producers, the majority (25.6%) belonged to CC8/ST239-MRSA clone. Our result revealed that about one-third of Panton-Valentine leukocidin (PVL)-positive strains belonged to CC30/ST30. The high-level mupirocin-resistant (HLMUPR) isolates belonged to CC8/ST239-MRSA (36%), CC30/ST30-MRSA (16%), CC8/ST8-MRSA (12%), CC5/ST5-MRSA (12%), CC8/ST585-MRSA (8%), CC5/ST225-MRSA (8%), CC5/ST1637-MRSA (4%), and CC8/ST1465-MRSA (4%) lineages carrying mupA. The VRSA strain belonged to the CC8/ST239-MRSA, CC8/ST8-MRSA, and CC22/ST22-MRSA clonal lineages, carrying the vanA determinant. Conclusion: These findings highlight significant genotypic diversity and high biofilm formation among our isolates. From this study, we identified highly virulent strains of S. aureus associated with biofilm production and drug resistance; some of these strains were highly similar, highlighting the possibility of rapid spread. The high prevalence of CC8 and CC30 clones among S. aureus strains reflects the emergence of these lineages as successful clones in hospital wastewaters in Iran, which is a serious concern. The study highlights the importance of wastewater surveillance to understand genetic pattern and antimicrobial resistance profiles in surrounding communities, which can in turn support public health efforts.
Keywords: Staphylococcus aureus; hospitals; methicillin-resistant Staphylococcus aureus; vancomycin-resistant Staphylococcus aureus; wastewaters.
Copyright © 2025 Fatemeh Sadat Tabatabaie Poya et al. International Journal of Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Paulus G. K., Hornstra L. M., Alygizakis N., Slobodnik J., Thomaidis N., Medema G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. International Journal of Hygiene and Environmental Health . 2019;222(4):635–644. doi: 10.1016/j.ijheh.2019.01.004. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

